Team II Webserver Group
Members: Paarth Parekh, Shivam Sharma, Sooyoun Oh, Jayson Chao, Hanchen Wang
Introduction
Background
- Purpose:
- Investigate an unknown outbreak pathogen using raw genome sequence data from the Centers for Disease Control and Prevention (CDC) foodborne illness surveillance outbreak investigations
- Goal:
- Create a Predictive Web Server that automates the process of characterizing the Campylobacter jejuni and make recommendations for the outbreak control.
Objective
- Assemble the input reads
- Analyze the assembly and predict annotated genes
- Identifying the strain as a phylogenetic tree( or heatmap)
- Calculate distance from the strain in the existing database
- Virulence factor and antimicrobial resistance profiling
- Visualize results in an effective manner
Design Goals
- Mobile friendly
- Easy to use
- Minimal
Basic Pipeline Structure
This is the Basic Image for our Pipeline which describes the Input each part of the functionality takes in and the output.

Framework
DJANGO Back-end development connects the server side of our pipeline and database with the browser. We have used Django, a python web framework as it can integrate hardware at any level, and it can handle large amounts of traffic. It is also easy to implement and can enable the user to focus on the seperate functionality, without getting into the complexities of it.
Why Django?
- Compatibility with python code: Django easily incorporates backbone scripts from each other group.
- Database integration: Django has built-in support for many popular databases, while PHP must use outside packages to handle databases.
- Security: Django is more secure than PHP.
- Database accessibility: Django has an ORM system, which makes database manipulation easier than using SQL.
- Scalability: Django is designed for bigger projects than Flask.
- Community support: Django has a larger following, and it is easier to find troubleshooting support.
Front End
For Front end programming we have used:
- Bootstrap, a popular framework for building responsive websites
- HTML 5 doctype (the latest design and development standard)
- CSS stylesheet: style of website
- Javascript plugin support (jQuery): Alerts,Buttons, dropdowns, tooltips
Database
- Django provides connection to MySql, Sqlite, PostgreSQL.
- We’re using Sqlite for our database, for its lightweight structure, and doesn’t need a heavy server (as in MySQL).
Features
Genome Assembly
- Performs de-novo assembly with FastQ files as input
- Runs the following tools:
- fastp: read pre-processing
- Spades: For Genome assembly
- The input FastQ files must be paired-end reads
- For Information on the tools visit: Team2_Genome_Assembly
- Outputs as FASTA file
- Visualisation: Quast output
Gene Prediction
- Gene finding in isolates assembled from Genome Assembly or user provided fasta file as input
- Runs the following tools:
- GeneMarkS-2 or Prodigal for CDS prediction
- Aragon for tRNA prediction
- Barrnap for rRNA prediction
- For more Information on tools visit: Team2_Gene_Prediction
- Outputs:
- For CDS: *.gff file, *.fna file, *.faa file
- For tRNA: *.fa file
- for rRNA: *.gff file, *.fa file
Website Architecture
- Server
- We’ll have used nginx in reverse proxy with gunicorn for our predictive web server.
- Gunicorn is appropriate for python based web applications and projects and directly interacts with our django project.
- Nginx sits on the outer layer and interacts directly with clients and manages security protocols.
- Nginx deals with large-sized files and manages the server load efficiently.
- Async Structure for Long Processes
- Celery (python) is an async task/job queue ideal for running long jobs in the background and update the user once the job is done. Celery can be integrated with Django and efficient error-handling can be performed as well.
- Email: We are using SendGrid as a cloud based platform to send emails to the user once their job is finished, using the wrappers in Django around the SNTP protocol.
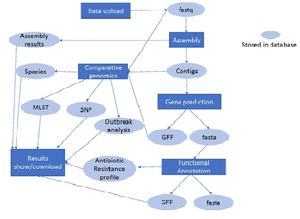
- Webpage Workflow
This is the entire workflow of our webpage with the blue indictor showing the parts of the pipeline stored in our database.
Access to Webserver
Here is Link to access our webserver: Cabunicrisis-Team2_webserver.
Here is our final presentation for Webserver: File:Team-2 Web Server Final.pdf
Reference
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics, btu170.
- Shifu Chen, Yanqing Zhou, Yaru Chen, Jia Gu; fastp: an ultra-fast all-in-one FASTQ preprocessor, Bioinformatics, Volume 34, Issue 17, 1 September 2018, Pages i884–i890, https://doi.org/10.1093/bioinformatics/bty560
- Philip Ewels, Måns Magnusson, Sverker Lundin and Max Käller. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics (2016) doi: 10.1093/bioinformatics/btw354 PMID: 27312411
- Bankevich, Anton et al. “SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.” Journal of computational biology : a journal of computational molecular cell biology vol. 19,5 (2012): 455-77. doi:10.1089/cmb.2012.0021
- ncbi.nlm.nih.gov/pmc/articles/PMC2952100/
- Gurevich, A., Saveliev, V., Vyahhi, N., & Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics, 29(8), 1072-1075.
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990) "Basic local alignment search tool." J. Mol. Biol. 215:403-410.
- Epps, S. V., Harvey, R. B., Hume, M. E., Phillips, T. D., Anderson, R. C., & Nisbet, D. J. (2013). Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. International journal of environmental research and public health, 10(12), 6292–6304. https://doi.org/10.3390/ijerph10126292
- Nucleotide BLAST: Search nucleotide databases using a nucleotide query. (n.d.). Retrieved from https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch
- Sheppard SK, Dallas JF, Wilson DJ, Strachan NJC, McCarthy ND, Jolley KA, et al. (2010) Evolution of an Agriculture-Associated Disease Causing Campylobacter coli Clade: Evidence from National Surveillance Data in Scotland. PLoS ONE 5(12): e15708. https://doi.org/10.1371/journal.pone.0015708
- Shuji Suzuki, Masanori Kakuta, Takashi Ishida, Yutaka Akiyama, Faster sequence homology searches by clustering subsequences, Bioinformatics, Volume 31, Issue 8, 15 April 2015, Pages 1183–1190, https://doi.org/10.1093/bioinformatics/btu780