Team II Gene Prediction Group
Team 2: Gene Prediction
Team Members: Danielle Temples, Kara Keun Lee, Paarth Parekh, Shuting Lin
Introduction
Our Project
Purpose: Investigate an unknown outbreak pathogen using raw genome sequence data from the Centers for Disease Control and Prevention (CDC) foodborne illness surveillance outbreak investigations
Overall Objective: Identify and characterize the pathogenic organism, make recommendations for the outbreak control, and build a public webserver that automates the computational steps
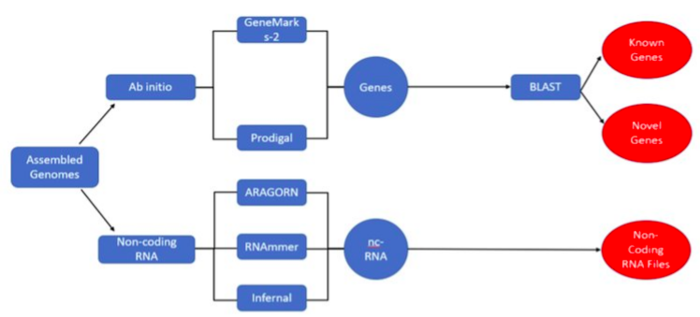
Our Objective: From assembled genomes, predict genes or features using different prediction methods and evaluate selected tools on their accuracy and performance
What is Gene Prediction?
Identification of the regions of genomic DNA that encode genes, which are fragments of DNA that encodes a functional molecule:
- Protein-coding genes
- RNA genes
- May also include other functional elements (i.e. regulatory regions)
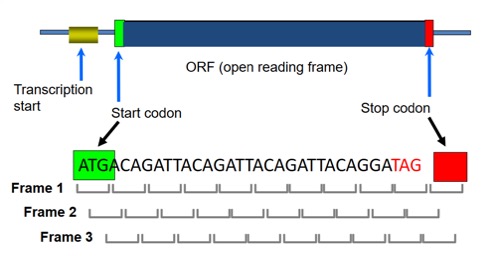
Prokaryotic Genome
Prokaryotic Genomes have a high gene density and do not contain introns in their protein-coding regions. Genes are called Open Reading Frames or “ORFs” (include start & stop codon). Prediction of prokaryotic genes tends to be relatively simpler with contiguous ORFs. However, overlapping ORFs and short genes can cause issues. Each gene is an ORF, but not every ORF is a gene.
Homology Methods
- Makes predictions via comparisons with sequences of previously known genes
- Based on extrinsic information
- Can be used to validate/support Ab Initio findings
- Limited by the use of no new knowledge
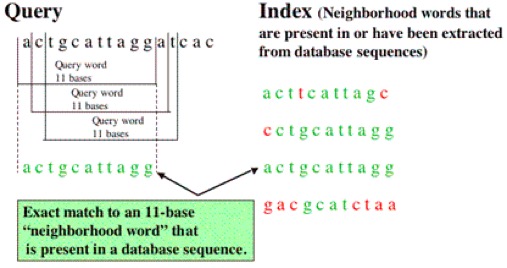
BLAST
Useful for dentifying species, locating domains, establishing phylogeny, DNA mapping, and comparison.
- Break query into words of length W
- Align words with sequence in database & identify matches
- Calculate T score for matches
- Extend sequence in both directions until score falls below cutoff (HSPs)
- Report hits that meet or exceed BLAST cutoff for statistically significant hits
Pros: 50 times faster than Dynamic Programming, allows for gapped matches
Cons: Less accurate than Smith-Waterman, may have low sensitivity
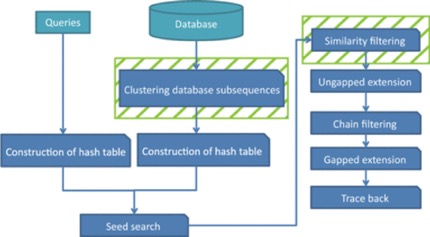
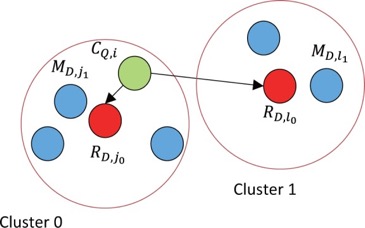
GHOSTZ
A new faster homology search method using database subsequence clustering.
- Sequences are extracted from a database & similar ones are clustered
- Construct into hash tables
- Use hash tables to select seeds for the alignments from representative sequences in the clusters
- Distance between a query subsequence and cluster representative is calculated
- Lower bounds calculated
- Similarity Filtering – if computed lower bound is less than or equal to distance threshold, continue
Pros: 200 times more efficient than BLAST, does not depend on search sensitivity
Cons: requires more memory usage
Database Selection
We obtained sequences from the RefSeq bacteria database since it is a BLAST accessible database that provides a "comprehensive, integrated, non-redundant, well-annotated set of sequences, including genomic DNA, transcripts, and proteins." It presents a stable reference for genome prediction as well as genome annotation which would assist the Functional Annotation group. Although we were fairly positive that our pathogen was Campylobacter jejuni, we chose to pull Campylobacter jejuni as well as Campylobacter coli data since these two are considered to be the most important enteropathogens among the Campylobacter spp. In addition, C. jejuni and C. coli have a strong relationship since they deviate from the same common ancestor.
Download Campylobacter jejuni and Campylobacter coli data from RefSeq bacteria database:
- 197 = taxid for Campylobacter jejuni
- 195 = taxid for Campylobacter coli
$ pip install ncbi-genome-download $ ncbi-genome-download --species-taxid 197 bacteria -F fasta -o Cjejuni_refseq_genomes --parallel 5 $ ncbi-genome-download --species-taxid 195 bacteria -F fasta -o Ccoli_refseq_genomes --parallel 5
Make BLAST custom database from all RefSeq .fna files :
$ makeblastdb -in <multifasta.fna> -dbtype nucl -out <mydb>
Ab Initio Methods
- Inspect the input sequence and searches for traces of gene presence
- Simplest method is to inspect ORFs
- Relies on probability models & specific DNA motifs (signals)
- Markov Models and Dynamic Programming
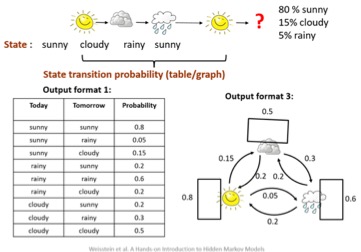
Hidden Markov Models
- Markov Model is a chain structured process where future states depend only on the present state
- Used to model randomly changing systems
- Hidden Markov Model (HMM) is a statistical Markov model with hidden states
- Viterbi Algorithm used to find the most likely sequence of hidden paths
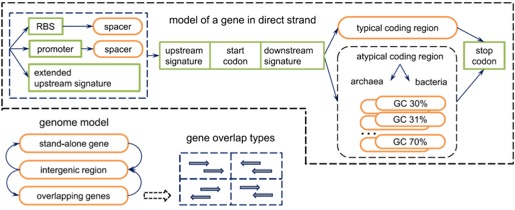
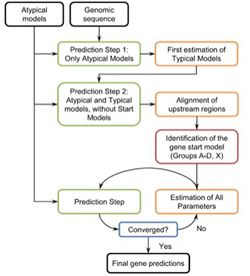
GeneMarkS-2
- Uses HMM and a self training algorithm (non supervised) to predict genes
- 5th Order HMM for coding and 2nd order for non-coding regions
- Identifies several different types of distinct sequence patterns
- The model which yields the highest log-odds score is selected
- Classifies the genome into 4 distinct groups:
- Group A: Typical Model of Prokaryotes having RBS sites having (SD)Consensus
- Group B: Atypical Model having RBS sites not having SD consensus
- Group C and D: Represent Bacterial and Archeal Genomes (Leaderless Transcription)
- Group X: Weak, Hard to classify regulatory signal patterns
- Stops after 10 iterations in the final prediction step, if it doesn’t converge
PRODIGAL
- Looks at GC bias for each of three codon positions and chooses the one with highest GC content
- Scores every start-stop pair above 90 bp in the entire genome based on simple GC codon statistics
- Penalizes or gives bonus to intergenic spaces according to gene distance
- Uses Dynamic Programming to force the program to choose between two heavily overlapping ORFS
Pros: runs unsupervised, handles gaps and partial genes, identifies translation initiation sites, predicts Genes in 3 formats (GFF/GenBank/Sequin)
Cons: sacrifices some genuine predictions to eliminate a much larger number of false identifications
Glimmer3
- Identifies genes within microbial DNA sequences (bacteria, archaea, and viruses)
- Uses Dynamic Programming to choose the highest-scoring set of ORFs and start sites
- Extracts every sufficiently long ORF from the sequence and scores it by the log-likelihood ratio of generating the ORF between trained models
- Uses Interpolated Markov Models (IMM) (combines 1st through 8th order Markov models)
- ORFs are scored from 3’ end to 5’ end, i.e., from stop codon back toward start codon, which helps find the start site
- For each ORF:
- Calculate the probability of the ORF sequence in each of the 6 possible reading frames
- If the highest-scoring frame corresponds to the reading frame of the ORF, mark the ORF as a gene
Cons: it does not work as well on high-GC genomes because it trains on long ORFs
Tool Evaluation
Homology
In order to choose the homology tool best suited for our pathogen, we decided to compare the GHOSTZ and BLAST results to SSEARCH results which uses Smith-Waterman algorithm. The more accurate homology tool would have the same subject sequence result as the subject sequence with the highest score in SSEARCH.
GHOSTZ requires specifically formatted database files for homology search, and these files can be generated from FASTA formatted DNA/protein sequence files. In Faster sequence homology searches by clustering subsequences, the authors generated a database from KEGG which contains 10 million protein sequences. We gathered 86,096 FASTA formatted Campylobacter jejuni/coli nucleotide sequences from the RefSeq bacterial genome database, so we did not foresee any problems with the database making step of GHOSTZ. However, after running for 40+ hours, we decided this was not an efficient use of our time allotted for this project. The authors do not detail the database making step in the paper and/or any potential issues, but this may be because the paper was published by GHOSTZ.
Ab Initio
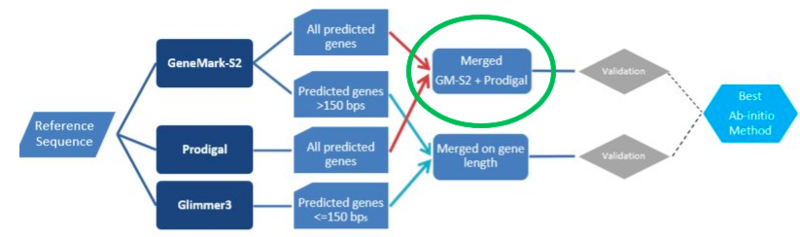
Tool Evaluation Plan:
- Use reference sequence on GeneMark-S2, PRODIGAL and Glimmer3
- Prepare MERGED data with bedtools in 2 ways:
- All predicted genes: PRODIGAL + GeneMark-S2
- Genes by gene length: >300 bps = GeneMark-S2; <=300 bps = Glimmer3
- Validate using “best” homology method (decided in pipeline above)
- Check for sensitivity, specificity, etc.
- Select the best Ab Initio method and proceed with our data
Testing Results:
Three Campylobacter jejuni reference genomes were used to evaluate tools and the combinations of tools. The predicted genes were blasted against a database made up of reference genome protein sequences and evaluated on the following parameters:
- TP = True Positive
- FP = False Positive
- FN = False Negative
- SE = TP / (TP+FN)
- PPV = TP / (TP+FP)
- Accuracy = TP / (TP+FP+FN)
SE was the highest for PRODIGAL + GMS-2 (99.4%) and marginally higher for combined prediction than single tool prediction, PPV was the highest for PRODIGAL (88.1%), and Accuracy was the highest for PRODIGAL + GMS-2 (86.0%).
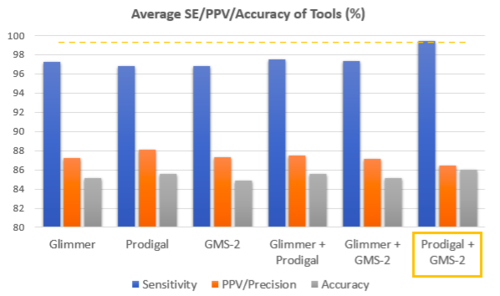
Tool Evaluation Results:
Based on our preliminary testing, merging GeneMarkS-2 and PRODIGAL predictions ("union") was selected as the best Ab-Initio method for our organism.
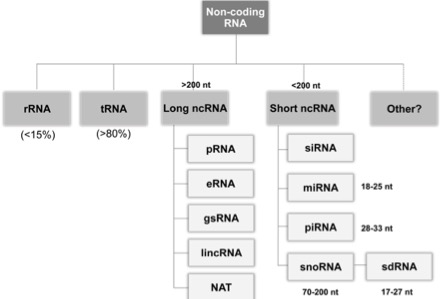
Non-Coding RNA Methods
- Non-coding RNA (ncRNA) is an RNA molecule that is not translated into a protein
- Transfer RNAs (tRNAs), ribosomal RNAs (rRNAs) and small RNAs(sRNAs)
- Protein synthesis/Translation (tRNA and rRNA) & gene regulation (sRNA)
- Related to antibiotic resistance
ARAGORN
- Homology based tool
- Uses heuristic algorithms that score the tRNA and tmRNA genes based on their sequence and secondary structure similarities
- An effective tRNA search program, with sensitivity better than other current heuristic tRNA search algorithms
RNAmmer
- Ab Initio based tool
- Uses Hidden Markov Models trained on data from 5s rRNA database
- Fast with little loss of sensitivity, enabling the analysis of a complete genome in less than a minute
- Location of rRNAs can be predicted with a very high accuracy
Infernal
- Implementation of covariance models (CMs)
- RNA homology search based on accelerated profile HMM methods and HMM-banded CM alignment methods
- 100-fold faster RNA homology searches and ∼10,000-fold acceleration over exhaustive non-filtered CM searches
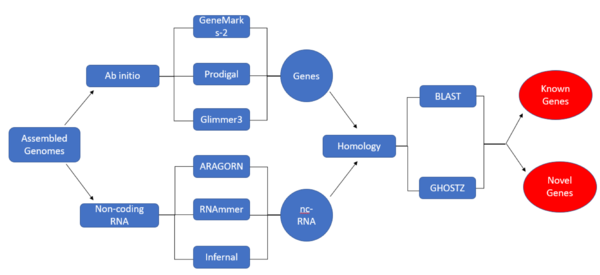
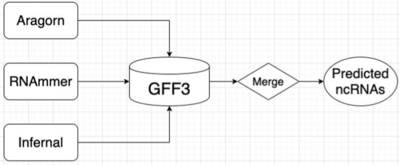
Initial Pipeline
Results
Ab-Initio
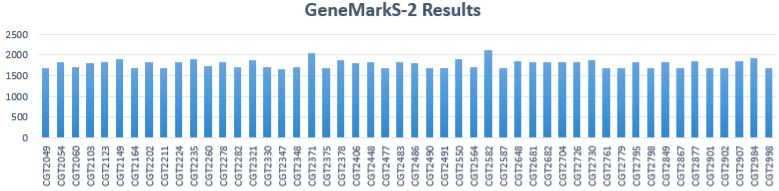
GeneMarkS-2 Results
$ gms2.pl --seq assembly_input --genome-type bacteria/auto --output output_gff –-format gff –-fnn output_nucleotide –-faa output_protein
- Total # of isolates: 50
- Average # of genes predicted per sample: 1780.9
- Minimum # of genes: 1668
- Maximum # of genes: 2107
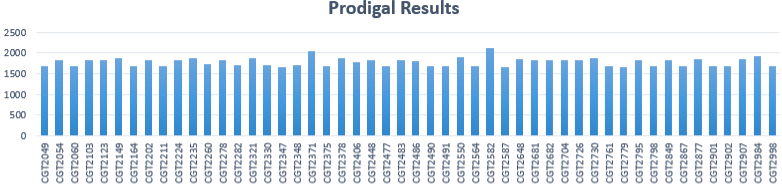
PRODIGAL Results
$ prodigal -i assembly_input –a output_protein –d output_nucleotide –o output_gff -f gff
- Total # of isolates: 50
- Average # of genes predicted per sample: 1778
- Minimum # of genes: 1660
- Maximum # of genes: 2105
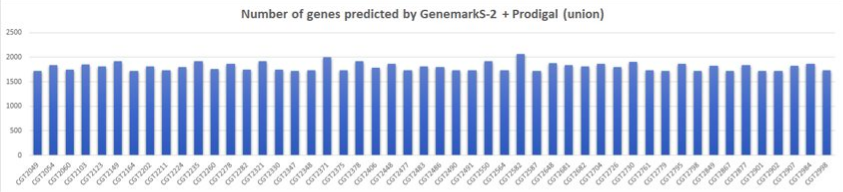
Union
Use bedtools intersect to get the union of GeneMarkS-2 results and PRODIGAL results:
# generate 3 parts of the union using bedtools intersect: sp.call(["bedtools","intersect","-a",gms_gff,"-b",prod_gff,"-wa","-v","-f","1.0","-r",">",intersect1_gff]) sp.call(["bedtools","intersect","-a",gms_gff,"-a",prod_gff,"-wa","-v","-f","1.0","-r",">",intersect2_gff]) sp.call(["bedtools","intersect","-a",gms_gff,"-b",prod_gff,"-f","1.0","-r",">",intersect3_gff])
Next, use bedtools getfasta to get the nucleotide sequence using the contigs files from the .gff files:
# generate 3 union fna files for blasting using union gff: sp.call(["samtools","faidx",contig_file]) sp.call(["bedtools","getfasta","-fi",contig_file,"-bed",union_gff,"-fo",union_fna])
Last, use transeq to get the union protein file from the union nucleotide file:
# generate 3 union faa files, maintain header format: sp.call(["transeq",union_fna,"-sformat","pearson","-trim","-clean","-outseq",union_faa]) sp.call(["sed","-i","'s/_[^_]*$//'",union_faa])
- Total # of isolates: 50
- Average # of genes predicted per sample: 1806 (95% CI: 1783,1829)
- Minimum # of genes: 1716
- Maximum # of genes: 2065
Homology
- Searches nucleotide databases using a nucleotide query
- Returns the most similar DNA sequences from the DNA database
- max_hsps = show the best HSP for every query-subject pair
- outfmt 6 = tabular format
- perc_identity 100 = only find exact matches
$ blastn -db <mydb> -query <contig> -out <filename> -max_hsps 1 –max_target_seqs 1 -outfmt 6 –perc_identity 100 –num_threads 5
Validation of Pathogen
Since our database was comprised of both Campylobacter jejuni and Campylobacter coli seqeunces, we were able to validate our pathogen with the BLASTn returned hits. The results showed a 70% alignment with C. jejuni versus a 30% alignment with C. coli.
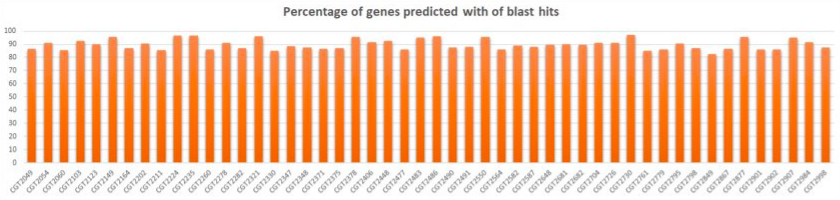
Validation of Ab-Initio Results
- Total # of isolates: 50
- Average # of BLAST hits per sample: 1623 (95% CI: 1587, 1659)
- Average % of genes with BLAST hits per sample: 89.8% (95% CI: 0.825, 0.972)
Mark genes in union_fna, union_faa, and union_gff files as KNOWN (K) or UNKNOWN (U) based on BLASTn hits (example):
Non-Coding RNA
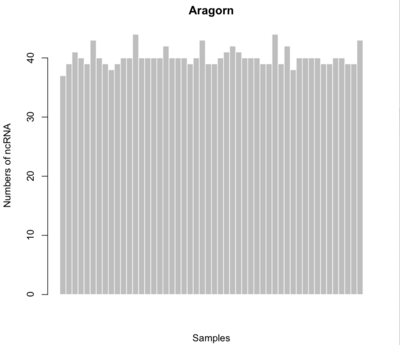
ARAGORN
$ aragorn -l -t -gc1 -w <input> -fo –o <output>
- Minimum # of tRNAs: 37
- Maximum # of tRNAs: 44
- Average # of tRNAS: 40
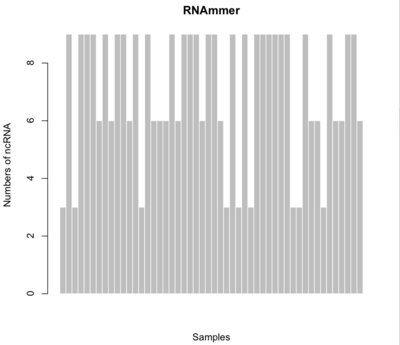
RNAmmer
$ ./rnammer -S bac -multi -gff rRNA.gff <input>
- Minimum # of rRNAs: 3
- Maximum # of rRNAs: 9
- Average # of rRNAS: 7.08
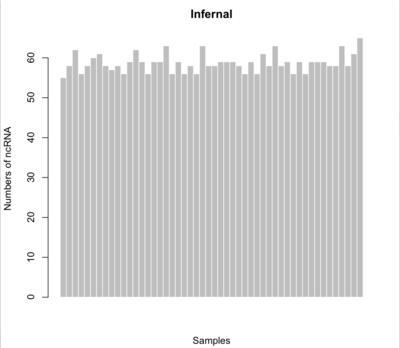
Infernal
$ cmscan -E 1e-06 --rfam --tblout <tblout_output> --noali --fmt 2 --clanin Rfam12.2.claninfo Rfam.cm <input>
- Minimum # of ncRNAs: 55
- Maximum # of ncRNAs: 65
- Average # of ncRNAS: 58.72
Convert & Merge ncRNAs
- Convert all formats from different tools into GFF3 format
- Deduplicate and take the union of all the predicted ncRNA results
Final Pipeline
References
About RefSeq. (n.d.). Retrieved from https://www.ncbi.nlm.nih.gov/refseq/about/
Epps, S. V., Harvey, R. B., Hume, M. E., Phillips, T. D., Anderson, R. C., & Nisbet, D. J. (2013). Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. International journal of environmental research and public health, 10(12), 6292–6304. https://doi.org/10.3390/ijerph10126292
Nucleotide BLAST: Search nucleotide databases using a nucleotide query. (n.d.). Retrieved from https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch
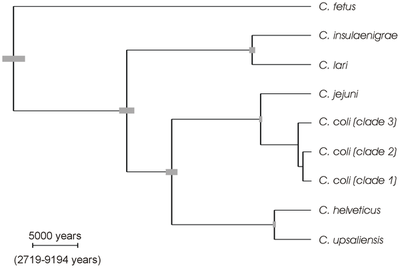
Sheppard SK, Dallas JF, Wilson DJ, Strachan NJC, McCarthy ND, Jolley KA, et al. (2010) Evolution of an Agriculture-Associated Disease Causing Campylobacter coli Clade: Evidence from National Surveillance Data in Scotland. PLoS ONE 5(12): e15708. https://doi.org/10.1371/journal.pone.0015708
Shuji Suzuki, Masanori Kakuta, Takashi Ishida, Yutaka Akiyama, Faster sequence homology searches by clustering subsequences, Bioinformatics, Volume 31, Issue 8, 15 April 2015, Pages 1183–1190, https://doi.org/10.1093/bioinformatics/btu780