Team III Genome Assembly Group: Difference between revisions
mNo edit summary |
No edit summary |
||
| Line 7: | Line 7: | ||
== Introduction == | == Introduction == | ||
Genome assembly is the process of combining the reads generated by sequencing technology into contiguous sequences that ultimately span a large portion of a genome. This can either be accomplished by examining how the reads overlap or examining their similarity to a reference genome. A standard genome assembly pipeline includes quality control, trimming, post-trimming quality control, sequence assembly, and assembly validation [1]. The objective of this project was to assemble bacterial genomes from 50 fastq files provided by CDC PulseNET. All of the samples were generated by paired-end sequencing on the Illumina platform. | |||
== Methods == | == Methods == | ||
| Line 16: | Line 14: | ||
[[File:3_assembly_pipeline.png|border|center|700px]] | [[File:3_assembly_pipeline.png|border|center|700px]] | ||
<center>'''Figure 1:''' Proposed assembly pipeline. | <center>'''Figure 1:''' Proposed assembly pipeline. fastp and MultiQC are used for quality control/trimming, sequence assembling is done in SPAdes, and assembly validation is conducted in Quast and BUSCO. The yellow bubbles represent the output files generated at each step.</center> | ||
=== Quality Control/Trimming === | === Quality Control/Trimming === | ||
| Line 33: | Line 31: | ||
[[File:3_QC_pretrim.jpg|border|center|600px]] | [[File:3_QC_pretrim.jpg|border|center|600px]] | ||
<center>'''Figure 2:''' Pre-trim quality control curves of our 50 samples. Plot generated using fastp and MultiQC.</center> | <center>'''Figure 2:''' Pre-trim quality control curves of our 50 samples. Plot generated using fastp and MultiQC. A) Read 1 quality curves; B) Read 2 quality curves.</center> | ||
Read 1 had high quality reads, so each of the 50 read 1 samples passed the mean quality score threshold of 28. However, read 2 was of lower quality, with only 25 samples passing the same mean quality threshold. This was mostly due to low quality regions at the 5' end of read 2. Because of this notable difference in quality between the two reads, we decided to trim read 1 and read 2 using different parameters. According to | Read 1 had high quality reads, so each of the 50 read 1 samples passed the mean quality score threshold of 28. However, read 2 was of lower quality, with only 25 samples passing the same mean quality threshold. This was mostly due to low quality regions at the 5' end of read 2. Because of this notable difference in quality between the two reads, we decided to trim read 1 and read 2 using different parameters. According to a comparison of trimming parameters on the assembly quality of ''Cyanoderma ruficeps'' genomes, trimming does not have a clear effect on assembly quality but speeds up most assembly implementations [2]. Therefore, in order to obtain high quality data to pass to the assemblers, we decided to completely trim the low-quality regions at the 3' and 5' ends of each read while retaining the high-quality middle regions. | ||
==== Trimming Parameters ==== | ==== Trimming Parameters ==== | ||
| Line 41: | Line 39: | ||
The following arguments were supplied to fastp in order to trim our data: <code>-f 5 -F 30 -t 10 -e 28 -c -5 3 -M 27.</code> | The following arguments were supplied to fastp in order to trim our data: <code>-f 5 -F 30 -t 10 -e 28 -c -5 3 -M 27.</code> | ||
* -f 5 - globally | * -f 5 - globally trims 5 bases from 5' end of mate 1 | ||
* -F 30 - globally | * -F 30 - globally trims 30 bases from 5' end of mate 2 | ||
* -t 10 - | * -t 10 - globally trims 10 bases from 3' end of both mates | ||
* -e 28 - | * -e 28 - discards reads with an average quality score under 28 | ||
* -c - turns on paired-end base correction, which slightly increased the quality of the 3' end of mate 2 | * -c - turns on paired-end base correction, which slightly increased the quality of the 3' end of mate 2 | ||
* -5 3 - turns on sliding window trimming from 5' end | * -5 3 - turns on sliding window trimming from 5' end with a window size of 3 | ||
* -M 27 - quality threshold of 27 for sliding window | * -M 27 - sets a quality threshold of 27 for sliding window | ||
=== Assembly === | === Assembly === | ||
| Line 62: | Line 60: | ||
===== MaSuRCA ===== | ===== MaSuRCA ===== | ||
'''Version:''' 3.3.5 | |||
MaSuRCA, or the Maryland Super-Read Celera Assembler, uses a combination of the OLC and de Bruijn Graph algorithms to assemble data. MaSuRCA normally creates longer contigs and offers higher N50 values. It works best on untrimmed data, which is possibly responsible for its slower computational time [2]. | |||
===== SPAdes ===== | ===== SPAdes ===== | ||
| Line 74: | Line 76: | ||
[[File:3_QC_posttrim.jpg|border|center|600px]] | [[File:3_QC_posttrim.jpg|border|center|600px]] | ||
<center>'''Figure 3''': Post-trim quality control curves of our 50 samples | <center>'''Figure 3''': Post-trim quality control curves of our 50 samples. Plot generated using fastp and MultiQC. A) Read 1 quality curves; B) Read 2 quality curves.</center> | ||
After trimming, the quality of our read 1 data improved slightly. However, the largest difference can be observed in read 2 data, with all 50 of the samples passing the mean quality threshold of a Phred score of 28. The 5' and 3' ends of some of the read 2 samples are of low quality, but more aggressive trimming would most probably also eliminate high-confidence regions. | |||
[[File:3_filtered_reads.png|border|center|600px]] | [[File:3_filtered_reads.png|border|center|600px]] | ||
<center>'''Figure 4''': Percentage of reads in each sample that passed the filters, generated using fastp and MultiQC.</center> | <center>'''Figure 4''': Percentage of reads in each sample that passed the filters, generated using fastp and MultiQC.</center> | ||
A vast majority of reads in each sample passed our filter, with at most 11% of reads falling below our quality and length thresholds and at most []% of bases trimmed from each sample. | |||
=== Post-Assembly Validation === | === Post-Assembly Validation === | ||
| Line 86: | Line 91: | ||
==== BUSCO ==== | ==== BUSCO ==== | ||
= References = | == Conclusions== | ||
== References == | |||
# Dominguez Del Angel V, Hjerde E, Sterck L et al. Ten steps to get started in Genome Assembly and Annotation [version 1; peer review: 2 approved]. ''F1000Research'' 2018, '''7'''(ELIXIR):148 (https://doi.org/10.12688/f1000research.13598.1) | |||
# Yang, S.-F.; Lu, C.-W.; Yao, C.-T.; Hung, C.-M. To Trim or Not to Trim: Effects of Read Trimming on the De Novo Genome Assembly of a Widespread East Asian Passerine, the Rufous-Capped Babbler (''Cyanoderma ruficeps'' Blyth). ''Genes'' 2019, '''10''', 737. | |||
Revision as of 21:39, 24 February 2020
Group Members: Deepali Kundnani, Aparna Maddala, Swetha Singu, Yiqiong Xiao, Ruize Yang
Lectures
Introduction
Genome assembly is the process of combining the reads generated by sequencing technology into contiguous sequences that ultimately span a large portion of a genome. This can either be accomplished by examining how the reads overlap or examining their similarity to a reference genome. A standard genome assembly pipeline includes quality control, trimming, post-trimming quality control, sequence assembly, and assembly validation [1]. The objective of this project was to assemble bacterial genomes from 50 fastq files provided by CDC PulseNET. All of the samples were generated by paired-end sequencing on the Illumina platform.
Methods
Pipeline for Testing
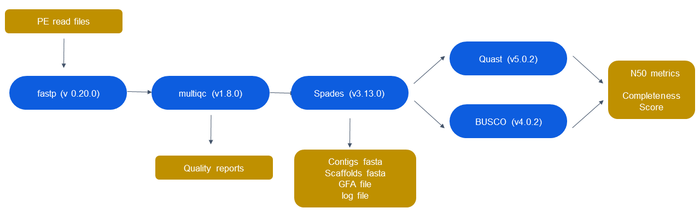
Quality Control/Trimming
Tools used:
- fastp (https://github.com/OpenGene/fastp)
- MultiQC (https://multiqc.info/)
Tool Selection
For quality control, we compared two tools, fastp and FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). We first proved that the two programs generated identical information when run on identical fastq files, after which we compared the information displayed in the reports for both. While FastQC creates highly informative per-base sequence quality graphs, it runs significantly slower than fastp. We were able to extract biologically significant information from the charts generated from fastp, so we decided to use fastp for quality control.
Afterwards, we compared fastp's trimming features with those of Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic). Our data did not contain adapters, so we did not need an additional tool like CutAdapt (https://cutadapt.readthedocs.io/en/stable/) to remove them. We showed that most, if not all, of Trimmomatic's trimming features can be replicated in fastp. Furthermore, fastp contains a feature specifically for paired-end data where it can use a high-read to correct low confidence bases in its mate. Fastp has the added advantage of combining both quality control and trimming into a single step, increasing the speed and usability of our pipeline. Since we had 50 input files in our pipeline, we used MultiQC to consolidate the 50 separate quality control reports generated by fastp into a single report.
Pre-Trim Quality Control
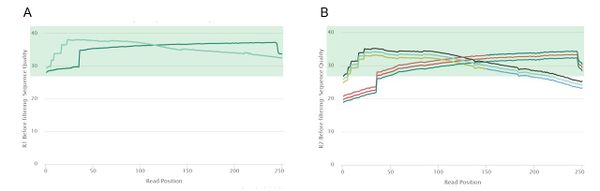
Read 1 had high quality reads, so each of the 50 read 1 samples passed the mean quality score threshold of 28. However, read 2 was of lower quality, with only 25 samples passing the same mean quality threshold. This was mostly due to low quality regions at the 5' end of read 2. Because of this notable difference in quality between the two reads, we decided to trim read 1 and read 2 using different parameters. According to a comparison of trimming parameters on the assembly quality of Cyanoderma ruficeps genomes, trimming does not have a clear effect on assembly quality but speeds up most assembly implementations [2]. Therefore, in order to obtain high quality data to pass to the assemblers, we decided to completely trim the low-quality regions at the 3' and 5' ends of each read while retaining the high-quality middle regions.
Trimming Parameters
The following arguments were supplied to fastp in order to trim our data: -f 5 -F 30 -t 10 -e 28 -c -5 3 -M 27.
- -f 5 - globally trims 5 bases from 5' end of mate 1
- -F 30 - globally trims 30 bases from 5' end of mate 2
- -t 10 - globally trims 10 bases from 3' end of both mates
- -e 28 - discards reads with an average quality score under 28
- -c - turns on paired-end base correction, which slightly increased the quality of the 3' end of mate 2
- -5 3 - turns on sliding window trimming from 5' end with a window size of 3
- -M 27 - sets a quality threshold of 27 for sliding window
Assembly
Reference-based vs. de novo assembly
Parameters considered when selecting assembly tool
[namedrop the two comparative studies we referenced here]
Tools considered
AbySS
MaSuRCA
Version: 3.3.5
MaSuRCA, or the Maryland Super-Read Celera Assembler, uses a combination of the OLC and de Bruijn Graph algorithms to assemble data. MaSuRCA normally creates longer contigs and offers higher N50 values. It works best on untrimmed data, which is possibly responsible for its slower computational time [2].
SPAdes
SKESA
Methods for testing assembly tools
Results
Post-Trimming Quality Control
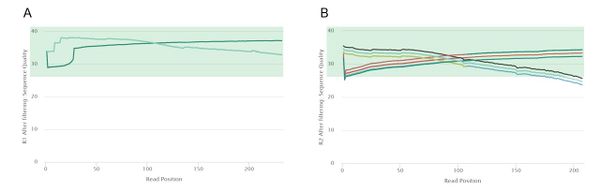
After trimming, the quality of our read 1 data improved slightly. However, the largest difference can be observed in read 2 data, with all 50 of the samples passing the mean quality threshold of a Phred score of 28. The 5' and 3' ends of some of the read 2 samples are of low quality, but more aggressive trimming would most probably also eliminate high-confidence regions.

A vast majority of reads in each sample passed our filter, with at most 11% of reads falling below our quality and length thresholds and at most []% of bases trimmed from each sample.
Post-Assembly Validation
quast
BUSCO
Conclusions
References
- Dominguez Del Angel V, Hjerde E, Sterck L et al. Ten steps to get started in Genome Assembly and Annotation [version 1; peer review: 2 approved]. F1000Research 2018, 7(ELIXIR):148 (https://doi.org/10.12688/f1000research.13598.1)
- Yang, S.-F.; Lu, C.-W.; Yao, C.-T.; Hung, C.-M. To Trim or Not to Trim: Effects of Read Trimming on the De Novo Genome Assembly of a Widespread East Asian Passerine, the Rufous-Capped Babbler (Cyanoderma ruficeps Blyth). Genes 2019, 10, 737.