Team II Genome Assembly Group: Difference between revisions
| Line 3: | Line 3: | ||
=== Introduction === | === Introduction === | ||
=== | === In-Class Presentations === | ||
[[File: Presentation 1.pdf]] | [[File: Presentation 1.pdf]] | ||
Revision as of 15:42, 29 January 2020
Team 2 Genome Assembly
Introduction
In-Class Presentations
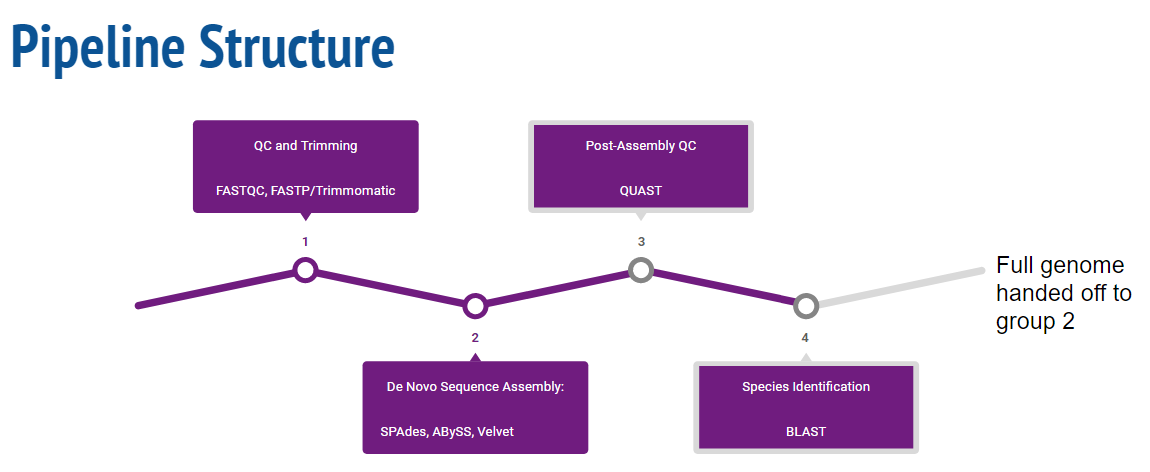
Pipeline Overview
Quality Control and Trimming
de Novo Assembly
Post-Assembly QC
Species Identification
Works Cited
Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics, btu170. Shifu Chen, Yanqing Zhou, Yaru Chen, Jia Gu; fastp: an ultra-fast all-in-one FASTQ preprocessor, Bioinformatics, Volume 34, Issue 17, 1 September 2018, Pages i884–i890, https://doi.org/10.1093/bioinformatics/bty560 Philip Ewels, Måns Magnusson, Sverker Lundin and Max Käller. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics (2016) doi: 10.1093/bioinformatics/btw354 PMID: 27312411 Bankevich, Anton et al. “SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.” Journal of computational biology : a journal of computational molecular cell biology vol. 19,5 (2012): 455-77. doi:10.1089/cmb.2012.0021 https://www.melbournebioinformatics.org.au/tutorials/tutorials/assembly/assembly-protocol/ https://bpa-csiro-workshops.github.io/btp-manuals-md/modules/btp-module-velvet/velvet/ ncbi.nlm.nih.gov/pmc/articles/PMC2952100/ Gurevich, A., Saveliev, V., Vyahhi, N., & Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics, 29(8), 1072-1075. Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990) "Basic local alignment search tool." J. Mol. Biol. 215:403-410.