Team I Comparative Genomics Group: Difference between revisions
Lmckinney8 (talk | contribs) |
Lmckinney8 (talk | contribs) |
||
| Line 36: | Line 36: | ||
''E. coli'' Mobile Genetic Elements | ''E. coli'' Mobile Genetic Elements | ||
* Bacterial cells transfer DNA between one another in three distinct ways: | * Bacterial cells transfer DNA between one another in three distinct ways (see Figure 3): | ||
** Transduction (1) | ** Transduction (1) | ||
** Conjugation (2) | ** Conjugation (2) | ||
Revision as of 17:54, 12 April 2020
Team 1 Comparative Genomics
Team members: Heather Patrick, Lawrence McKinney, Laura Mora, Manasa Vegesna, Kenji Gerhardt, Hira Anis
Introduction and Objectives
Comparative genomics is a field in biomedical research in which the genomic features of different organisms are compared. In short, it involves the comparison of one genome to another. This type of comparative analysis can be utilized to discover what lies hidden within the sequences of genomes by comparing sequencing information. Comparative genomics has utilities in gene prediction, regulatory element prediction, phylogenomics, pharmacogenomics, pathogenicity and more. For the purposes of our analysis, we will employ comparative genomics tools to conduct an outbreak analysis. More specifically, we will compare bacterial genomes generated from Illumina next-generation sequence data to generate knowledge that will help us identify and characterize a bacterial outbreak strain of Escherichia coli (E. coli). Will will then apply our computational results to known biological insights and matched epidemiological to further characterize the identified bacterial strain. This data will be used to propose containment and treatment options that can be used by public health professional to address the foodbourne illness.
Our Data:
- 50 isolates of Escherichia coli from an outbreak of foodborne illnesses. The genomes have been assembled and fully annotated.
- Epidemiological data consisting of: times, locations (states), and ingested foods of each case.
Our Bacteria:
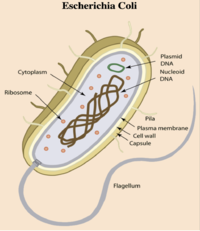
- E. coli is a gram-negative bacterium composed of numerous strains and serotypes (see Figure 1).
- E. coli contains plasmids (mobile genetic elements ) which generate genome diversity by promoting homologous recombination, horizontal gene transfer between bacteria, and can confer antimicrobial resistance and virulence.
- About ~46% of E. coli genome is conserved among all strains (core genome)
- E. coli occurs naturally in the lower part of the intestines of humans and warm-blooded animals, and under certain conditions, even commensal, “nonpathogenic” strains can cause infection.
- E. coli is typically transmitted through ingestion of contaminated food and water, person-to-person contact, contact with fomites.
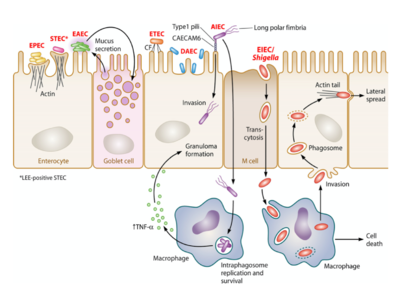
- There are 8 types of pathogenic strains of E. coli (see Figure 2):
- Enteropathogenic E. coli(EPEC)
- Enteroaggregative E. coli (EAEC)
- Enterotoxigenic E. coli (ETEC)
- Enteroinvasive E. coli (EIEC)
- Enterohamerrhagic E. coli (EHEC)
- Diffusely Adherent E. coli (DAEC)
- Adherent Invasive E. coli (AIEC)
- Shiga Toxin (Stx) producing Enteroaggregative E. coli (STEAEC)
- Strains representative of a pathotype contained shared genes as well as unique genes.
- Pathogenic E. coli is typically transmitted through ingestion of contaminated food and water, person-to-person contact, or contact with fomites. It typically invades and colonizes in the epithelium of the intestines.
E. coli Mobile Genetic Elements
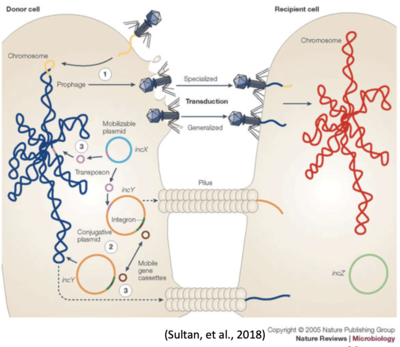
- Bacterial cells transfer DNA between one another in three distinct ways (see Figure 3):
- Transduction (1)
- Conjugation (2)
- Transformation (3)
- Transduction and conjugation depend on mobile genetic elements (MGEs), including most large plasmids and some bacteriophages.
- Pathogenomic analysis of the numerous plasmids present within representative strains of E. coli pathotypes (and commensal E. coli) has revealed considerable diversity and plasticity within these MGEs.
- Plasmids and bacteriophages play a major role in generating genome diversity by promoting homologous recombination and horizontal gene transfer between bacteria.
Team Objectives
- Compare and contrast functional & structural features of isolates.
- Antibiotic Resistance profile
- Virulence profile
- Differentiate outbreak vs. sporadic strains.
- Characterize the virulence and antibiotic resistance functional features of outbreak isolates.
- Identify the source and spread of the outbreak.
- Recommend outbreak response and treatment.
Methods
Results
Conclusion
In-Class Presentations
- Comparative Genomics Background and Strategy:
- Comparative Genomics Final Results:
References
- Chen X, Zhang Y, Zhang Z, Zhao Y, Sun C, Yang M, Wang J, Liu Q, Zhang B, Chen M, Yu J, Wu J, Jin Z and Xiao J (2018) PGAweb: A Web Server for Bacterial Pan-Genome Analysis. Front. Microbiol. 9:1910. doi: 10.3389/fmicb.2018.01910
- Maiden MC, Jansen van Rensburg MJ, Bray JE, et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol. 2013;11(10):728-36.
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, et al. (2018) MUMmer4: A fast and versatile genome alignment system. PLOS Computational Biology 14(1): e1005944. https://doi.org/10.1371/journal.pcbi.1005944
- Perez-Losada M, Arenas M, Castro-Nallar E. Microbial sequence typing in the genomic era. Infection, Genetics and Evolution. 2018;63:346-359. http://dx.doi.org/10.1016/j.meegid.2017.09.022
- Strockbine N, Bopp C, Fields P, Kaper J, Nataro J. 2015. Escherichia, Shigella, and Salmonella, p 685-713. In Jorgensen J, Pfaller M, Carroll K, Funke G, Landry M, Richter S, Warnock D (ed), Manual of Clinical Microbiology, Eleventh Edition. ASM Press, Washington, DC. doi: 10.1128/9781555817381.ch37
- Sultan, I., Rahman, S., Jan, A. T., Siddiqui, M. T., Mondal, A. H., & Haq, Q. M. R. (2018). Antibiotics, Resistome and Resistance Mechanisms: A Bacterial Perspective. Frontiers in Microbiology, 9(2066). doi:10.3389/fmicb.2018.02066
- Trees E, Rota P, Maccannell D, Gerner-smidt P.. Molecular Epidemiology, p 131-159. In Jorgensen J, Pfaller M, Carroll K, Funke G, Landry M, Richter S, Warnock D (ed), Manual of Clinical Microbiology, Eleventh Edition. ASM Press, Washington, DC. 2015. doi: 10.1128/9781555817381.ch10