Team II Genome Assembly Group: Difference between revisions
| Line 24: | Line 24: | ||
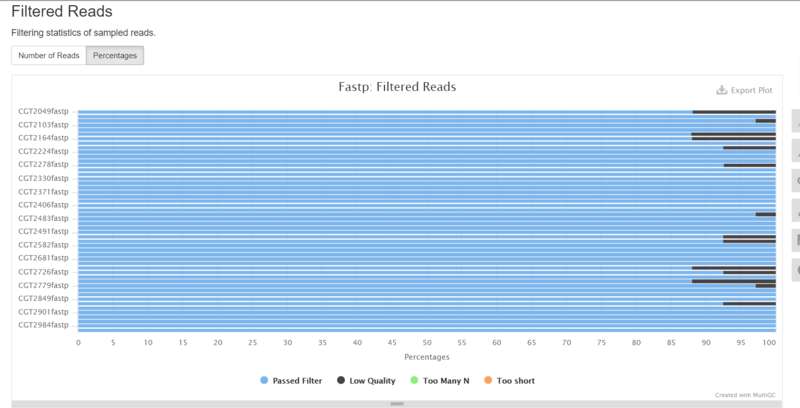
The 50 output html files were then combined using MultiQC (https://multiqc.info/) and the following parameters were found to optimize post-trim quality for all 50 inputs. | The 50 output html files were then combined using MultiQC (https://multiqc.info/) and the following parameters were found to optimize post-trim quality for all 50 inputs. | ||
[[File: filtered_reads.PNG|border| | [[File: filtered_reads.PNG|border|800px]] | ||
=== de Novo Assembly === | === de Novo Assembly === | ||
Revision as of 17:01, 3 February 2020
Team 2 Genome Assembly
Introduction
Taking the input of 50 paired fastq files from an unknown species, genome assembly and species identification were performed by trimming and aligning these short DNA sequences.
In-Class Presentations
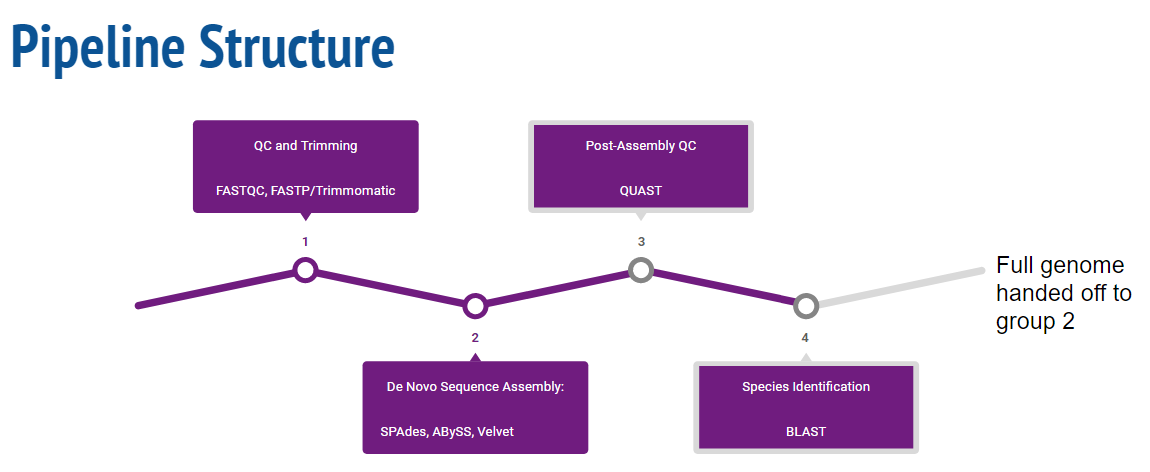
Pipeline Overview
Quality Control and Trimming
Before de Novo assembly can begin, input fastq files needed to be evaluated and processed for quality. Subsequently, low quality reads as well as adapters required trimming to ensure a more accurate genome assembly. The tool used for pre-trimming quality control was FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Following installation, the 100 provided fastq files were called into FastQC and the data summary text files locations were recorded. Summary files were iterated over and per-base sequence quality was compared to a threshold of 30. The first position to pass the threshold was selected to be the headcropping point for trimming. Subsequently, the final position to pass the quality control parameters was selected to be the tail cropping point for trimming. Trimming was then completed by FastP (https://github.com/OpenGene/fastp).
Following the generation of FastQC reports, output html files were aggregated using MultiQC (https://multiqc.info/). The following charts summarize the pre-QC fastq files.
Before FastP was selected as a tool for trimming, it was compared to Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic), another widely used trimming tool. FastP was found to be significantly faster than Trimmomatic, and also offered two major features that when implemented, optimized the complexity and runtime of this process. Firstly, FastP offers automatic adapter sensing, and since 50 paired end reads were passed through the pipeline, this feature allowed adapter trimmimg to be highly customized per fastq sequence. Secondly, output files from calling FastP include its own post-trim QC html and json files--negating the need for a subsequent calling of FastQC.
The 50 output html files were then combined using MultiQC (https://multiqc.info/) and the following parameters were found to optimize post-trim quality for all 50 inputs.
de Novo Assembly
Post-Assembly QC
The general rule of a high-quality assembly is the low number and long length of assembled contigs, in other words, larger N50 value.
QUAST (QUality ASsessment Tool) (http://quast.sourceforge.net/index.html) could evaluate the quality of the assembled contigs generated from various assemblers with full range of metrics. Among various publicly available tools for post-assembly QC, QUAST was selected based on several advantages. Firstly and most importantly, QUAST allows the usage without the reference genome, which satisfies the objective of the current project. Furthermore, instead of comparing a single N50 value, QUAST outputs multiple Nx (where 0<x<100) values to give better insight into the overall variation in contig length. Lastly, the various types of visualization reports would facilitate comparing the results from several assemblers.
Species Identification
Species identification could be achieved by extracting SSU fragments from the assembled contigs and searching for matches in the available public databases with taxonomies (e.g. SILVA, RDP, and Greengenes).
The other simple method would be aligning the assembled contigs against the NCBI non-redundant (nr) database which could be achieved on the NCBI BLAST web page (https://blast.ncbi.nlm.nih.gov/Blast.cgi). BLAST (Basic Local Alignment Search Tool) detects the local alignments based on k-mer search. Among various BLAST tools, the BLASTn (nucleotide BLAST) is applicable for comparing the nucleotide query sequence against the nucleotide nr database.
Works Cited
Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics, btu170. Shifu Chen, Yanqing Zhou, Yaru Chen, Jia Gu; fastp: an ultra-fast all-in-one FASTQ preprocessor, Bioinformatics, Volume 34, Issue 17, 1 September 2018, Pages i884–i890, https://doi.org/10.1093/bioinformatics/bty560 Philip Ewels, Måns Magnusson, Sverker Lundin and Max Käller. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics (2016) doi: 10.1093/bioinformatics/btw354 PMID: 27312411 Bankevich, Anton et al. “SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.” Journal of computational biology : a journal of computational molecular cell biology vol. 19,5 (2012): 455-77. doi:10.1089/cmb.2012.0021 https://www.melbournebioinformatics.org.au/tutorials/tutorials/assembly/assembly-protocol/ https://bpa-csiro-workshops.github.io/btp-manuals-md/modules/btp-module-velvet/velvet/ ncbi.nlm.nih.gov/pmc/articles/PMC2952100/ Gurevich, A., Saveliev, V., Vyahhi, N., & Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics, 29(8), 1072-1075. Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990) "Basic local alignment search tool." J. Mol. Biol. 215:403-410.