Team II Genome Assembly Group: Difference between revisions
| Line 11: | Line 11: | ||
=== Quality Control and Trimming === | === Quality Control and Trimming === | ||
Before de Novo assembly can begin, input fastq files needed to be evaluated and processed for quality. Subsequently, low quality reads as well as adapters required trimming to ensure a more accurate genome assembly. The tool used for pre-trimming quality control was FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). | |||
=== de Novo Assembly === | === de Novo Assembly === | ||
Revision as of 15:44, 29 January 2020
Team 2 Genome Assembly
Introduction
In-Class Presentations
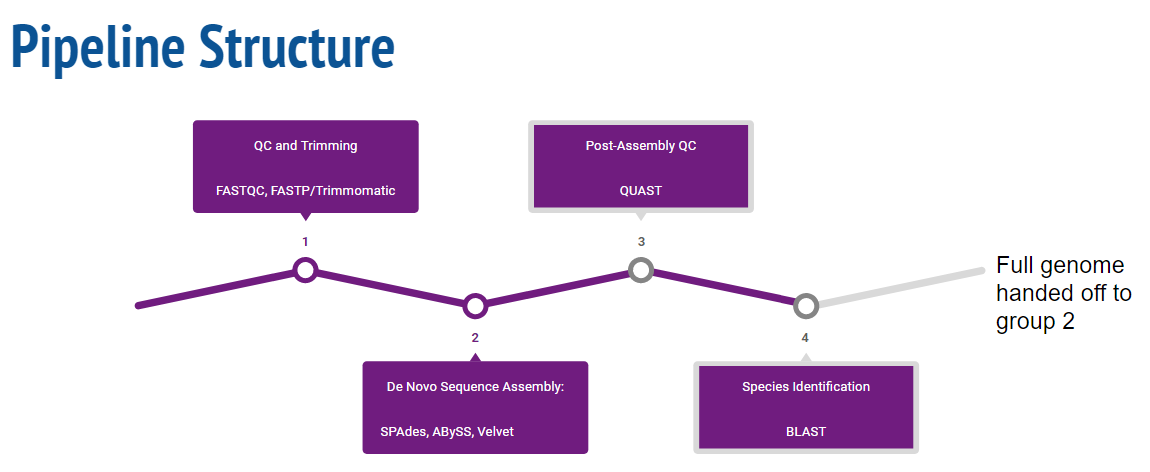
Pipeline Overview
Quality Control and Trimming
Before de Novo assembly can begin, input fastq files needed to be evaluated and processed for quality. Subsequently, low quality reads as well as adapters required trimming to ensure a more accurate genome assembly. The tool used for pre-trimming quality control was FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
de Novo Assembly
Post-Assembly QC
Species Identification
Works Cited
Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics, btu170. Shifu Chen, Yanqing Zhou, Yaru Chen, Jia Gu; fastp: an ultra-fast all-in-one FASTQ preprocessor, Bioinformatics, Volume 34, Issue 17, 1 September 2018, Pages i884–i890, https://doi.org/10.1093/bioinformatics/bty560 Philip Ewels, Måns Magnusson, Sverker Lundin and Max Käller. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics (2016) doi: 10.1093/bioinformatics/btw354 PMID: 27312411 Bankevich, Anton et al. “SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.” Journal of computational biology : a journal of computational molecular cell biology vol. 19,5 (2012): 455-77. doi:10.1089/cmb.2012.0021 https://www.melbournebioinformatics.org.au/tutorials/tutorials/assembly/assembly-protocol/ https://bpa-csiro-workshops.github.io/btp-manuals-md/modules/btp-module-velvet/velvet/ ncbi.nlm.nih.gov/pmc/articles/PMC2952100/ Gurevich, A., Saveliev, V., Vyahhi, N., & Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics, 29(8), 1072-1075. Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990) "Basic local alignment search tool." J. Mol. Biol. 215:403-410.