Team III Gene Prediction Group: Difference between revisions
No edit summary |
No edit summary |
||
| Line 11: | Line 11: | ||
=='''Initial Pipeline'''== | =='''Initial Pipeline'''== | ||
[[File: 2.PNG]] | |||
=='''Coding homology - Tools'''== | |||
===[https://blast.ncbi.nlm.nih.gov/Blast.cgi '''BLAST''']=== | |||
BLAST (Basic Local Alignment Search Tool) finds regions of similarity between two sequences by locating matches between them. Instead of comparing every residue against each other, BLAST uses short "word" segments to create alignment "seeds." which reduces the search space. BLAST then extends the alignment in both directions according to a threshold set by the user. | |||
===[http://hmmer.org/ '''HMMER''']=== | |||
HMMER is used for searching sequence databases for sequence homologs, and for making sequence alignments, using Hidden Markov Models (HMMs). It makes a profile HMM of the query that assigns a position-specific scoring system for substitution, insertions, and deletions. A profile HMM is a variant of an HMM relating specifically to biological sequences. Profile HMMs turn a multiple sequence alignment into a position-specific scoring system, which can be used to align sequences and search databases for remotely homologous sequences. It provides a formal probabilistic framework for sequence comparison and improves detection of remote homology by (i) enabling position-specific residue and gap scoring based on a query profile (ii) calculating the signal of homology-based on the more powerful ‘Forward/Backward’ HMM algorithm that computes not just one best-scoring alignment, but a sum of support over all possible alignments. | |||
===[https://uni-tuebingen.de/fakultaeten/mathematisch-naturwissenschaftliche-fakultaet/fachbereiche/informatik/lehrstuehle/algorithms-in-bioinformatics/software/diamond/ '''DIAMOND''']=== | |||
<Add description here> | |||
=='''Non-Coding homology - Tools'''== | =='''Non-Coding homology - Tools'''== | ||
Revision as of 01:44, 8 March 2020
Introduction
Gene prediction is the process of identifying the regions of genomic DNA that encode genes which primarily include protein-coding and non-coding genes. Gene prediction is an important process that aids in the identification of fundamental and essential elements of the genome.
With the overall goal to investigate a foodborne outbreak caused by a prokaryotic organism, our team developed a pipeline for the prediction of coding and non-coding genes in prokaryotes.
Our final objective was to carry out a thorough and exhaustive prediction of all coding and non-coding genes of the 50 assembled genomes provided by the Genome Assembly team.
In prokaryotic genomes, DNA sequences that encode proteins are transcribed into mRNA, and then RNA is usually translated directly into proteins without significant modification. They have a higher gene density in comparison to eukaryotes.

There are two gene prediction methods - Homology methods and ab-initio methods. Homology-based gene prediction methods rely on extrinsic evidence and makes predictions by comparing with sequences of previously known genes. Homology-based approaches are helpful as they are used to validate ab-initio methods but are limited by existing knowledge, sometimes computationally expensive, and require an extensive and vast database as all genes are not expressed at the same time. Ab-initio gene prediction methods, on the other hand, rely on intrinsic evidence of the genome. These methods detect promoter sequences, start and stop codons and GC content to predict ORFs. The disadvantages of ab-initio methods are the high false-positive rate, no experimental verification and not robust as homology-based tools to name a few. The main algorithms used in various ab-initio tools are Hidden Markov Modelling (HMM), Interpolated Markov Modelling (IMM) and Dynamic Programming.
Initial Pipeline
Coding homology - Tools
BLAST
BLAST (Basic Local Alignment Search Tool) finds regions of similarity between two sequences by locating matches between them. Instead of comparing every residue against each other, BLAST uses short "word" segments to create alignment "seeds." which reduces the search space. BLAST then extends the alignment in both directions according to a threshold set by the user.
HMMER
HMMER is used for searching sequence databases for sequence homologs, and for making sequence alignments, using Hidden Markov Models (HMMs). It makes a profile HMM of the query that assigns a position-specific scoring system for substitution, insertions, and deletions. A profile HMM is a variant of an HMM relating specifically to biological sequences. Profile HMMs turn a multiple sequence alignment into a position-specific scoring system, which can be used to align sequences and search databases for remotely homologous sequences. It provides a formal probabilistic framework for sequence comparison and improves detection of remote homology by (i) enabling position-specific residue and gap scoring based on a query profile (ii) calculating the signal of homology-based on the more powerful ‘Forward/Backward’ HMM algorithm that computes not just one best-scoring alignment, but a sum of support over all possible alignments.
DIAMOND
<Add description here>
Non-Coding homology - Tools
ARAGORN
Aragorn is a computer program identifies tRNA and tmRNA genes. The program employs heuristic algorithms to predict tRNA secondary structure, based on homology with recognized tRNA consensus sequences and ability to form a base-paired cloverleaf. tmRNA genes are identified using a modified version of the BRUCE program.
Infernal
Infernal is for searching DNA sequence databases for RNA structure and sequence similarities. It is an implementation of a special case of profile stochastic context-free grammars called covariance models (CMs). A CM is like a sequence profile, but it scores a combination of sequence consensus and RNA secondary structure consensus, so in many cases, it is more capable of identifying RNA homologs that conserve their secondary structure more than their primary sequence.
Non-Coding Ab initio - Tools
RNAmmer
RNAmmer predicts ribosomal RNA genes in full genome sequences by utilizing two levels of Hidden Markov Models: An initial spotter model searches both strands. The spotter model is constructed from highly conserved loci within a structural alignment of known rRNA sequences.
Barrnap
Barrnap predicts the location of ribosomal RNA genes in genomes by using HMMER 3.1 for HMM searching in RNA:DNA style. It supports bacteria (5S,23S,16S), archaea (5S,5.8S,23S,16S), metazoan mitochondria (12S,16S) and eukaryotes (5S,5.8S,28S,18S).
Benchmarking of non-coding + Results inferred
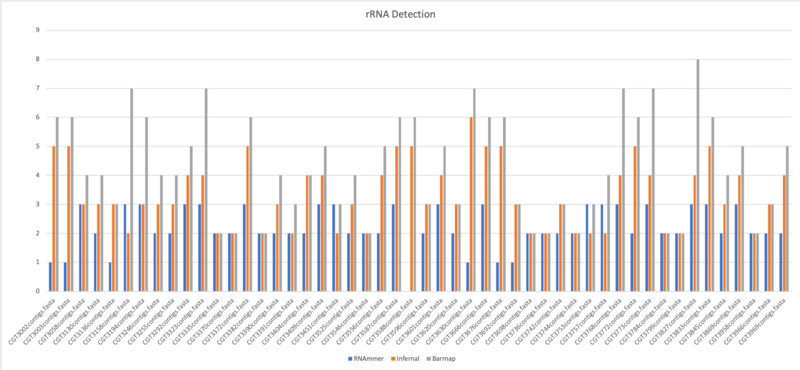
rRNA Comparison
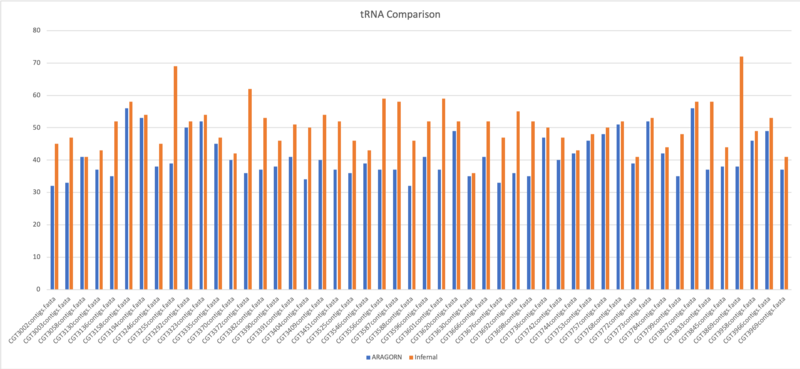
tRNA Comparison
Results: Non-Coding
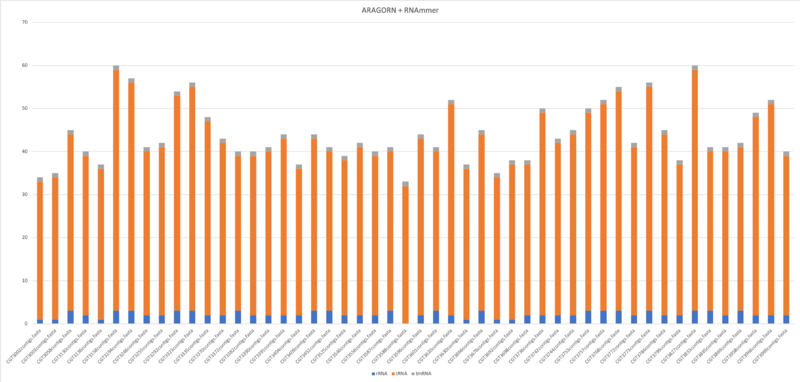
Aragorn & RNAmmer
$ aragorn -l -t -gc1 -w input.fasta -fo –o output.fasta $ aragorn -l -m -gc1 -w input.fasta -fo –o output.fasta
- Average of tRNA: 40.9
- Average of tmRNA: 1
- Average of rRNA: 2.2
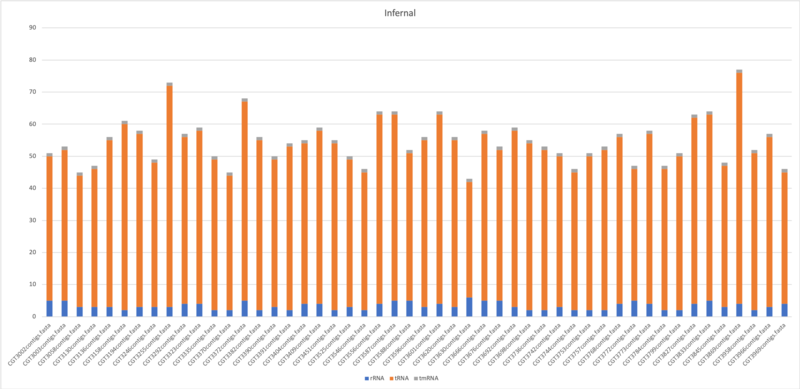
Infernal
$ cmscan --cut_ga --rfam --nohmmonly --tblout $output/$(basename $filename .fasta).tblout --fmt 2 --clanin Rfam.clanin Rfam.cm $filename > $output/$(basename $filename . fasta).cmscan
- Average of tRNA: 50.5
- Average of tmRNA: 1
- Average of rRNA: 3.34