Team III Gene Prediction Group
Group Members - Pallavi Misra, Sonali Gupta, Ahish Sujay, Jie Zhou, Cheng Sheng-Yi
Introduction
Gene prediction is the process of identifying the regions of genomic DNA that encode genes which primarily include protein-coding and non-coding genes. Gene prediction is an important process that aids in the identification of fundamental and essential elements of the genome.
With the overall goal to investigate a foodborne outbreak caused by a prokaryotic organism, our team developed a pipeline for the prediction of coding and non-coding genes in prokaryotes.
Our final objective was to carry out a thorough and exhaustive prediction of all coding and non-coding genes of the 50 assembled genomes provided by the Genome Assembly team.
In prokaryotic genomes, DNA sequences that encode proteins are transcribed into mRNA, and then RNA is usually translated directly into proteins without significant modification. They have a higher gene density in comparison to eukaryotes.

There are two gene prediction methods - Homology methods and ab-initio methods. Homology-based gene prediction methods rely on extrinsic evidence and makes predictions by comparing with sequences of previously known genes. Homology-based approaches are helpful as they are used to validate ab-initio methods but are limited by existing knowledge, sometimes computationally expensive, and require an extensive and vast database as all genes are not expressed at the same time. Ab-initio gene prediction methods, on the other hand, rely on intrinsic evidence of the genome. These methods detect promoter sequences, start and stop codons and GC content to predict ORFs. The disadvantages of ab-initio methods are the high false-positive rate, no experimental verification and not robust as homology-based tools to name a few. The main algorithms used in various ab-initio tools are Hidden Markov Modelling (HMM), Interpolated Markov Modelling (IMM) and Dynamic Programming.
Initial Pipeline
Coding homology - Tools
BLAST
BLAST (Basic Local Alignment Search Tool) finds regions of similarity between two sequences by locating matches between them. Instead of comparing every residue against each other, BLAST uses short "word" segments to create alignment "seeds." which reduces the search space. BLAST then extends the alignment in both directions according to a threshold set by the user.
HMMER
HMMER is used for searching sequence databases for sequence homologs, and for making sequence alignments, using Hidden Markov Models (HMMs). It makes a profile HMM of the query that assigns a position-specific scoring system for substitution, insertions, and deletions. A profile HMM is a variant of an HMM relating specifically to biological sequences. Profile HMMs turn a multiple sequence alignment into a position-specific scoring system, which can be used to align sequences and search databases for remotely homologous sequences. It provides a formal probabilistic framework for sequence comparison and improves detection of remote homology by (i) enabling position-specific residue and gap scoring based on a query profile (ii) calculating the signal of homology-based on the more powerful ‘Forward/Backward’ HMM algorithm that computes not just one best-scoring alignment, but a sum of support over all possible alignments.
DIAMOND
Like BLAST, DIAMOND uses seed-and-extend paradigm. The algorithm first searches for matches of seeds (short stretches of the query sequence) in the reference database, and this is followed by an 'extend' phase that aims to compute a full alignment. What lends DIAMOND its speed is the use of double indexing, an approach that determines the list of all seeds and their locations in both the query and reference sequences. DIAMOND also optimizes its storing of seeds in memory efficient way.
Coding ab-initio - Tools
PRODIGAL
PRODIGAL (PROkaryotic DYnamic programming Gene-finding ALgorithm) scores individual ORFs using various features and scoring rules, and then performs dynamic programming on all pairs of start-and-stop triplets to find the maximum scoring path. The adopted features Prodigal includes are GC bias in first, second, and third positions of each codon, frequency of hexamers, ORF length, upstream sequence resembling ORF, etc. The connection of a start node to its corresponding stop node represents a gene, whereas the connection of a 3' end to a new 5' end represents intergenic space.
GLIMMER
GLIMMER(Gene Locator and Interpolated Markov ModelER) searches for long-ORFs and generates a training data set to which it trains all six Markov models of coding and non-coding DNA from zero to eight order. After calculating the probabilities from the above data, GLIMMER decides to either use fixed order Markov model or interpolated Markov model. Performed by program “build-imm”. a.If the no. observation > 400 = Fixed order Markov model b.If the no. observation < 400 = Interpolated Markov model Glimmer obtains score for every long-ORF generated and if score is greater than a certain threshold, GLIMMER predicts it as a gene. Performed by program “glimmer”.
GeneMarkS-2
It uses a model derived by self-training for finding species-specific (native) genes. It uses precomputed heuristic models designed to identify harder-to-detect genes, horizontally transferred genes, etc.
Benchmarking of coding + Results inferred
Workflow:
Evaluation Metrics:
Sensitivity: (True Positive)/(True Positive + False Negative)
False Discovery Rate: (False Positive)/(True Positive + False Positive)
Precision: (True Positive)/(True Positive + False Positive)
Sensitivity Result:
False Discovery Rate (FDR) Result:
Precision Result:
Inference:
False Discovery Rate for Glimmer is highly fluctuating. Hence we did not include Glimmer in our final pipeline. <elaborate>
Non-Coding homology - Tools
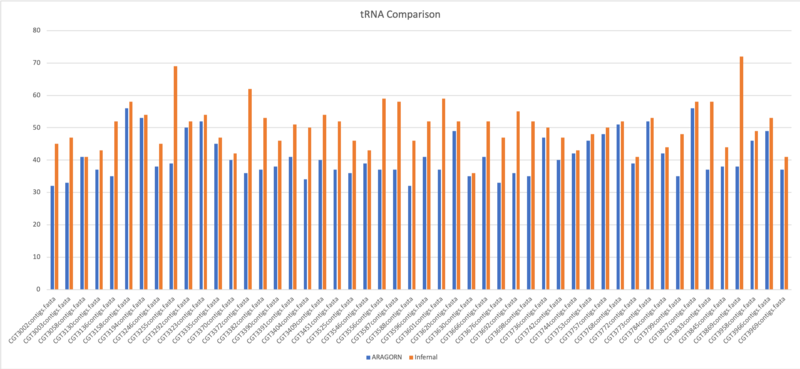
ARAGORN
Aragorn is a computer program identifies tRNA and tmRNA genes. The program employs heuristic algorithms to predict tRNA secondary structure, based on homology with recognized tRNA consensus sequences and ability to form a base-paired cloverleaf. tmRNA genes are identified using a modified version of the BRUCE program.
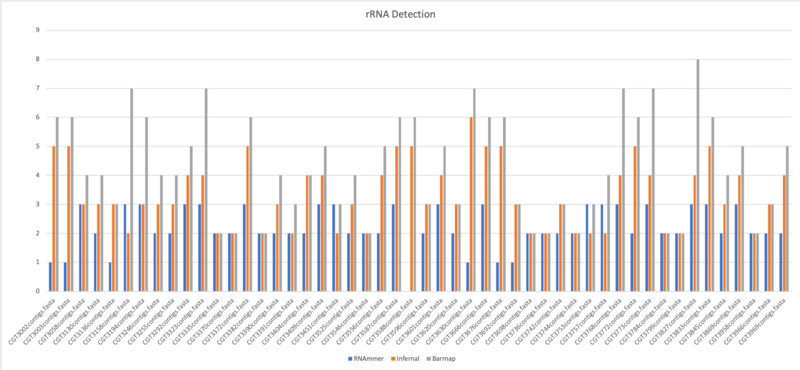
Infernal
Infernal is for searching DNA sequence databases for RNA structure and sequence similarities. It is an implementation of a special case of profile stochastic context-free grammars called covariance models (CMs). A CM is like a sequence profile, but it scores a combination of sequence consensus and RNA secondary structure consensus, so in many cases, it is more capable of identifying RNA homologs that conserve their secondary structure more than their primary sequence.
Non-Coding ab-initio - Tools
RNAmmer
RNAmmer predicts ribosomal RNA genes in full genome sequences by utilizing two levels of Hidden Markov Models: An initial spotter model searches both strands. The spotter model is constructed from highly conserved loci within a structural alignment of known rRNA sequences.
Barrnap
Barrnap predicts the location of ribosomal RNA genes in genomes by using HMMER 3.1 for HMM searching in RNA:DNA style. It supports bacteria (5S,23S,16S), archaea (5S,5.8S,23S,16S), metazoan mitochondria (12S,16S) and eukaryotes (5S,5.8S,28S,18S).
Benchmarking of non-coding + Results inferred
rRNA Comparison
tRNA Comparison
Final Pipeline
Results: Coding
Prodigal
$ prodigal -i contig_file -d fasta_file -f gff -o gff_file
GeneMarkS-2
$ perl gms2.pl --seq contig_file --genome-type bacteria -fnn nucl_file --output gff_file --formal gff
Merging results of ab-initio tools
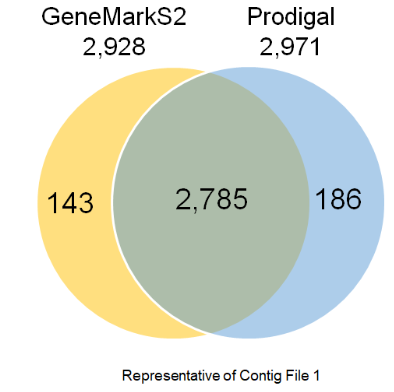
To find reciprocal overlap between Prodigal and GeneMarkS2
$ bedtools intersect -f 1.0 -r -a gms2.out -b prodigal.out > Intersection
To find the sequences which are only in GeneMarkS2
$ bedtools intersect -f 1.0 -v -r -a gms2.out -b prodigal.out > GeneMark_only
To find the sequences which are only in Prodigal
$ bedtools intersect -f 1.0 -v -r -a prodigal.out -b gms2.out > Prodigal_only
Combine these three files- concatenate Input this to blast to get TP and FP
Final output:
GFF TP
Fasta TP
GFF FP
Fasta FP
Results: Non-Coding
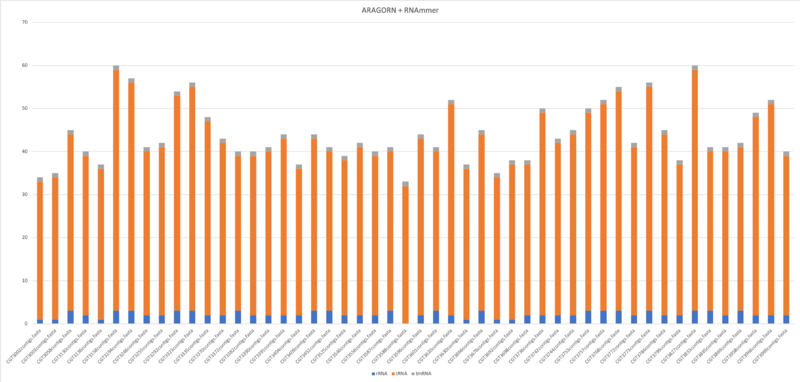
Aragorn & RNAmmer
$ aragorn -l -t -gc1 -w input.fasta -fo –o output.fasta $ aragorn -l -m -gc1 -w input.fasta -fo –o output.fasta
- Average of tRNA: 40.9
- Average of tmRNA: 1
- Average of rRNA: 2.2
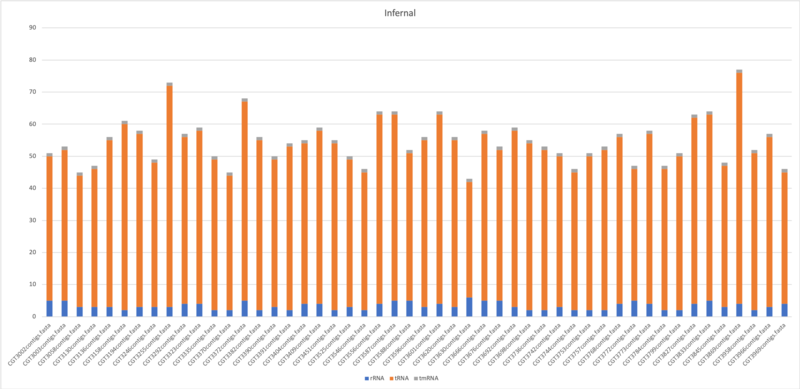
Infernal
$ cmscan --cut_ga --rfam --nohmmonly --tblout $output/$(basename $filename .fasta).tblout --fmt 2 --clanin Rfam.clanin Rfam.cm $filename > $output/$(basename $filename . fasta).cmscan
- Average of tRNA: 50.5
- Average of tmRNA: 1
- Average of rRNA: 3.34
References
https://www.researchgate.net/figure/Panel-A-shows-the-structure-of-a-prokaryotic-gene-The-protein-coding-region-is-flanked_fig1_267784124 [accessed 7 Mar, 2020]
Goodswen SJ, Kennedy PJ, Ellis JT. Evaluating high-throughput ab initio gene finders to discover proteins encoded in eukaryotic pathogen genomes missed by laboratory techniques.
PLoS One. 2012;7(11):e50609. doi: 10.1371/journal.pone.0050609. Epub 2012 Nov 30. PubMed PMID: 23226328; PubMed Central PMCID: PMC3511556.
Angelova, Mihaela & Kalajdziski, Slobodan & Kocarev, Ljupco. (2010). Computational Methods for Gene Finding in Prokaryotes. ICT Innovations. 1. 1857-7288
Birney E, Durbin R. Using GeneWise in the Drosophila annotation experiment. Genome Res. 2000;10(4):547–548. doi:10.1101/gr.10.4.547
Stephen F. Altschul, Warren Gish, Webb Miller, Eugene W. Myers, David J. Lipman, Basic local alignment search tool, Journal of Molecular Biology,Volume 215, Issue 3
Skewes-Cox P, Sharpton TJ, Pollard KS, DeRisi JL (2014) Profile Hidden Markov Models for the Detection of Viruses within Metagenomic Sequence Data. PLoS ONE 9(8): e105067.
https://doi.org/10.1371/journal.pone.0105067
Lomsadze, Alexandre, et al. "Modeling leaderless transcription and atypical genes results in more accurate gene prediction in prokaryotes." Genome research 28.7 (2018): 1079-1089.
Altschul, Stephen F., et al. "Basic local alignment search tool." Journal of molecular biology 215.3 (1990): 403-410.
Finn, Robert D., Jody Clements, and Sean R. Eddy. "HMMER web server: interactive sequence similarity searching." Nucleic acids research 39.suppl_2 (2011): W29-W37.
Buchfink, Benjamin, Chao Xie, and Daniel H. Huson. "Fast and sensitive protein alignment using DIAMOND." Nature methods 12.1 (2015): 59.
Hyatt, Doug, et al. "Prodigal: prokaryotic gene recognition and translation initiation site identification." BMC bioinformatics 11.1 (2010): 119.
Salzberg, Steven L., et al. "Microbial gene identification using interpolated Markov models." Nucleic acids research 26.2 (1998): 544-548.
Lomsadze, Alexandre, et al. "Modeling leaderless transcription and atypical genes results in more accurate gene prediction in prokaryotes." Genome research 28.7 (2018): 1079-1089.
Laslett, Dean, and Bjorn Canback. "ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences." Nucleic acids research 32.1 (2004): 11-16.
Nawrocki, Eric P., Diana L. Kolbe, and Sean R. Eddy. "Infernal 1.0: inference of RNA alignments." Bioinformatics 25.10 (2009): 1335-1337.
Lagesen, Karin, et al. "RNAmmer: consistent and rapid annotation of ribosomal RNA genes." Nucleic acids research 35.9 (2007): 3100-3108.