Team III Gene Prediction Group: Difference between revisions
| Line 16: | Line 16: | ||
==''' | =='''Results- non-Coding'''== | ||
==='''Aragorn & Barrnap'''=== | ==='''Aragorn & Barrnap'''=== | ||
$ aragorn -l -t -gc1 -w input.fasta -fo –o output.fasta | $ aragorn -l -t -gc1 -w input.fasta -fo –o output.fasta | ||
Revision as of 19:30, 7 March 2020
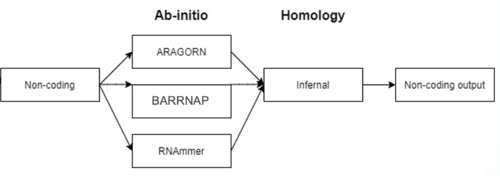
Non-Coding RNA
A non-coding RNA (ncRNA) is an molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.
ARAGORN
Aragorn is a computer program identifies tRNA and tmRNA genes. The program employs heuristic algorithms to predict tRNA secondary structure, based on homology with recognized tRNA consensus sequences and ability to form a base-paired cloverleaf. tmRNA genes are identified using a modified version of the BRUCE program.
Barrnap
Barrnap predicts the location of ribosomal RNA genes in genomes by using HMMER 3.1 for HMM searching in RNA:DNA style. It supports bacteria (5S,23S,16S), archaea (5S,5.8S,23S,16S), metazoan mitochondria (12S,16S) and eukaryotes (5S,5.8S,28S,18S).
RNAmmer
RNAmmer predicts ribosomal RNA genes in full genome sequences by utilizing two levels of Hidden Markov Models: An initial spotter model searches both strands. The spotter model is constructed from highly conserved loci within a structural alignment of known rRNA sequences.
Infernal
Infernal is for searching DNA sequence databases for RNA structure and sequence similarities. It is an implementation of a special case of profile stochastic context-free grammars called covariance models (CMs). A CM is like a sequence profile, but it scores a combination of sequence consensus and RNA secondary structure consensus, so in many cases, it is more capable of identifying RNA homologs that conserve their secondary structure more than their primary sequence.
Results- non-Coding
Aragorn & Barrnap
$ aragorn -l -t -gc1 -w input.fasta -fo –o output.fasta $ aragorn -l -m -gc1 -w input.fasta -fo –o output.fasta
- Average of tRNA: 40.9
- Average of tmRNA: 1
- Average of rRNA: 2.2
Infernal
- Average of tRNA: 50.5
- Average of tmRNA: 1
- Average of rRNA: 3.34