Team I Gene Prediction Group
Members: Maria Ahmad, Hira Anis, Jessica Mulligan, Priya Narayanan, Aaron Pfennig, Winnie Zheng
Introduction
Prokaryotic Gene Feature
Figure 1. General Prokaryotic gene structure
- Prokaryotic genes have a relatively well-understood promoter sequence, such as a regulatory sequence, which can regulate the transcription of the gene into an mRNA.
- Each prokaryotic gene has open reading frames (ORF) which start with start codons and end with end codons with no interruptions (end-codons) in-between, so it can provide a good, but not assured prediction of the protein-coding regions.
Gene Prediction
Gene prediction or gene finding is a process of identifying the regions of genomic DNA that encode genes. It is devised two-classes of methods that use similarity-based (homology) searches and ab-initio prediction to capture the compositional differences among coding regions which will be translated into protein and non-coding DNA which can be translated into tRNAs and rRNAs.
Project Goal
The main goal of our project is to finish the gene prediction for the assembled gene from E.coli given by the genome assembly group.
Methods
Ab-initio Methods (CDS prediction)
- Features of Predicting protein-coding genes
- ORFs
- Signal Sensor: Regulatory motifs (RBS, SD, etc)
- Content Sensor: The codon usage bias, based on GC content, can help to distinguish coding sequence from surrounding non-coding sequence.
- Markov and Hidden Markov Model
Figure 2. State transition diagram of a hidden Markov model
- Markov model is a stochastic model that can model the dynamics of systems. This system is made up of known states with a known transition probability which depends only on current states. In other words, the future state depends only on the current state, not the previous one.
- Hidden Markov Model (HMM) is described as a Markov model of random changing systems that are made up of unobserved (hidden) states. This model can determine state transition probability which is the probability between each hidden state and generate observable nucleotides.
- GeneMarkS2
- Self-training algorithm based on an HMM.
- Models transcription domain to predict gene start more accurately.
- Includes a heuristic model designed to predict horizontally transferred genes.
- Pros and cons: GMS2 can have the highest sensitivity and specificity among different tools, but works on different gene regulatory motifs using learned and leaderless transcription.
./run_gms2.sh <path_to_genome>
- Glimmer3
- The Interpolated Markov Model(IMM) is at the core of Glimmer.
- Scoring the ORFs in reverse (stop codon back toward the start codon) relying on k-mer within the coding region.
- Trains on long ORFs.
- Pros and cons: Glimmer3 performs by most metrics but predicts the least short genes (<150 nt).
./run_glimmer3.sh <path_to_genome>
- Prodigal
- Identifying all ORFs and scores them using a dynamic programming approach.
- Refines predictions after training on the subset of ORFs.
- Pros and cons: Prodigals are trained on E.coli but predict the most genes start correctly in E.coil.
Non-Coding Gene Prediction
- Features of Non-Coding Gene Prediction
- Non-coding genes are RNAs that remain as transcripts and never get translated; their function lies in the RNA sequence and structure itself. Of the RNA types, two of the most prominent families are rRNA and tRNA. rRNA has structural functions as part of the ribosome and plays a part in translation, tRNAs are folded RNA structures which can ‘read’ codons from mRNA transcripts, and bring the correct amino acid to the growing peptide chain. There are different types of tRNA for different types of codons.
- tRNA
- To find the tRNAs in our organism, we used two tools: tRNAscan-SE and Aragorn. tRNAscan-SE is an industry-standard dating back to 1997. It uses covariance models and heuristic methods to find tRNAs. Aragorn is a newer tool and uses homology based methods to find tRNAs. It claims to be faster than tRNAscan-SE.
- tRNAscan-SE
- Homology based
- Averages ~40 seconds per isolate to run
- Predicts 73 tRNAs on average per isolate
- Aragorn
- Homology based
- Averages ~3.5 seconds per isolate to run
- Predicts 74 tRNAs on average per isolate
- 2D structural format visualization
- tRNAscan-SE
- To find the tRNAs in our organism, we used two tools: tRNAscan-SE and Aragorn. tRNAscan-SE is an industry-standard dating back to 1997. It uses covariance models and heuristic methods to find tRNAs. Aragorn is a newer tool and uses homology based methods to find tRNAs. It claims to be faster than tRNAscan-SE.
tRNAscan-SE ./tRNAscan-SE -B -L -H -o fileID.out -f fileID_structure.out -m fileID_stats.out -b fileID.bed -a fileID.fasta pathToIsolates Aragorn ./aragorn pathtoIsolates -o fileID.out -fo fileID.fasta -seq fileID_sequences -w fileID_batch
- rRNA
- To find the rRNAs in our organism, two tools were used: RNAmmer and Barrnap. RNAmmer is a program that uses hidden Markov models trained on data from the 5S ribosomal RNA database and the European ribosomal RNA database project. It manages to identify annotated and unannotated rRNAs. Barrnap is a newer tool, which has identified 75 rRNAs and used the latest version of HMMER (3.x.). Although Barrnap cites RNAmmer as more sophisticated and faster, Barrnap manages to outperform RNAmmer in our dataset.
- RNAmmer
- Homology based
- Predicts an average of 2 rRNAs per isolate.
- Output in gff format for each input contig file.
- rRNA identified : 5s,16s,23s in bacteria.
- Time taken: ~5 minutes per isolate.
- Pre-requisites : hmmer2.x, Perl.
- Relies on HMM (hmmer2.x) for speed and accuracy.
- Barrnap
- Homology based
- Predicts an average of 2 rRNAs per isolate.
- Output in gff format for each input contig file.
- rRNA identified : 5s,16s,23s in bacteria.
- Time taken: ~10 seconds per isolate.
- Multi-threading supported.
- Pre-requisites : Perl 5.xx (core modules only), nhmmer (part of HMMER 3.x), bedtools >= 2.27.0.
- RNAmmer
- To find the rRNAs in our organism, two tools were used: RNAmmer and Barrnap. RNAmmer is a program that uses hidden Markov models trained on data from the 5S ribosomal RNA database and the European ribosomal RNA database project. It manages to identify annotated and unannotated rRNAs. Barrnap is a newer tool, which has identified 75 rRNAs and used the latest version of HMMER (3.x.). Although Barrnap cites RNAmmer as more sophisticated and faster, Barrnap manages to outperform RNAmmer in our dataset.
RNAmmer ./rnammer -S bac -m tsu,lsu,ssu -gff <output_file> <input_file> Barrnap ./barrnap --outseq <output_file> --kingdom bac <input_file>
Tool Evaluation
Ab-initio Tools
- Use a reference gene, Escherichia_coli_cft073.ASM744v to test each tool by computing sensitivity (SN), positive predictive value (PPV) and specificity (SP) which only for start site prediction.
Calculation:
- SN = TP / (TP + FN)
- PPV = TP / (TP + FP)
- SP = TN / (TN + FP) (only start site prediction)
- TP=True Positive; TN=True Negative; FP= False Positive; FN=False Negative
Testing Results:
- The percentage of sensitivity, specificity and positive predictive value(only for the start site) were summarized on the following table. We also include the number of genes and running time in order to provide a stronger evaluation.
- GMS2 has higher sensitivity and start site positive predictive value. Prodigal has the shortest running time. At the same time, the Glimmer has the highest specificity.
Table 1. The performance evaluation for each tool by reference gene.
- On the 50 assembled genomes, comparing the predictions of GMS2, Prodigal, and Glimmer3 is to overlap each tool.
./get_overlaps.sh ./get_gene_counts.sh ./plot_venn_diagramm.py
Figure 3. The overlapping of GMS2, Prodigal, and Glimmer results.
Testing Results:
- Generally, the tools greatly overlap.
- Glimmer predicts several unique genes not predicted by the others.
- Prodigal and GMS2’s predictions have a large overlap.
Results
Ab-initio
- Performed a BLAST search of the predicted genes against SwissProt database
./make_diamond_db.sh ./run_diamond.sh
- Filtered BLAST results by >90% coverage of query to
./plot_comparision_blast_ab_intio.py
- Plot ratio of predicted genes with BLAST support
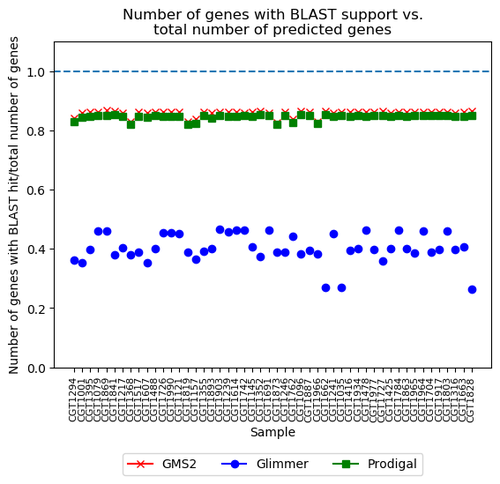
Figure 4. The ratio of each assembled genes with BLAST support from three tools.
Results: Prodigal and GMS2 perform more consistently, so the filtering does not influence the results.
- Plot bar plot of the number of predicted genes per sample
Figure 5. The number of predicted genes of each sample from GMS2, Prodigal, and Glimmer.
Results: The number of predicted genes between GMS2 and Prodigal is similar, but clearly, the Glimmer has a different number of predicted genes.
- Summary Results:
- Glimmer does not predict genes as consistently.
- Prodigal and GMS2 are very similar in terms of Sn/Sp and the number of predicted genes.
- But Prodigal runs ~13x faster.
Non-Coding Predictions
- tRNAscan-SE vs Aragon
Figure 6. The comparison between tRNAscan-SE results with Aragon results
Results: tRNAscan-SE and Aragon can predict almost the same number of tRNA genes. One is 3656 and another is 3709. Their identified tRNAs are also similar. One is 73 tRNAs on average and another is 74 tRNAs on average. However, Aragon is approx. 10x faster than tRNAscan-SE. Although Aragon does not output gff files, batch files can easily be converted to gff.
- RNAmmer vs Barrnap
Figure 7. The comparison between RNAmmer results with Barrnap results
Results:
- Both tools predict about the same number of rRNAs (48/50 of the contig files overlap)
- Differences in predictions in the samples:
- CGT1157_trim_assembled.fasta
- 23S_rRNA partial alignment (35%) - barrnap
- CGT1869_trim_assembled.fasta
- 5S_rRNA, 16S rRNA predictions for the sample - RNAmmer
- CGT1157_trim_assembled.fasta
- Barrnap < RNAmmer wrt. time taken
- Conclusion: barrnap for rRNA predictions (speed)
- DFAST framework: supports barrnap and RNAmmer
Pipeline
Initial Pipeline
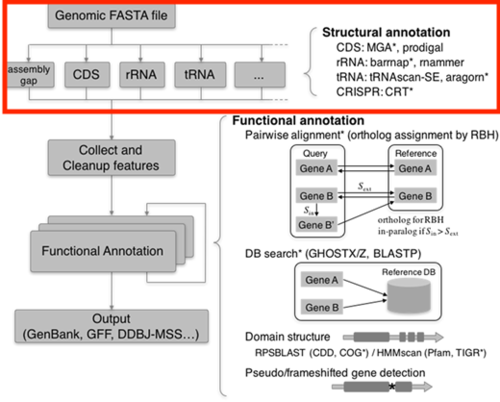
Through the research and previous paper, we first considered the pipeline as the figure above. We did the prediction for the genome based on as many tools as we can.
Final Pipeline
- Make use of the DFAST framework → prokaryotic annotation pipeline used by the DDBJ
- Runs all tools in parallel
- By default: Prodigal, Barrnarp, Aragorn
- Optionally: GMS2, RNAmmer, tRNAscan-SE
- → pipeline runs in ~20s per sample
- Output:
- GFF file
- Nucleotide seq of CDS (*_cds.fna)
- AA seq of CDS (*_protein.faa)
- Nucleotide seq of RNA (*_rna.fna)
Reference
[1] Prokaryotic Gene Structure. Wikipedia. https://en.wikipedia.org/wiki/Template:Prokaryote_gene_structure
[2] Wang, Z., Chen, Y., & Li, Y. (2004). A brief review of computational gene prediction methods. Genomics, proteomics & bioinformatics, 2(4), 216–221. doi:10.1016/s1672-0229(04)02028-5
[3] Toledo, Tomer & Katz, Romina. (2009). State Dependence in Lane-Changing Models. Transportation Research Record. 2124. 81-88. 10.3141/2124-08.
[4] Lomsadze Alexandre. et al. (2017) Improved prokaryotic gene prediction yields insights into transcription and translation mechanisms on whole genome scale. 1–24. doi: 10.1101/193490.
[5] Lomsadze, Alexandre et al. (2017) “GeneMarkS-2 : Raising Standards of Accuracy in Gene Recognition.” .
[6] A.L. Delcher, K.A. Bratke, E.C. Powers, and S.L. Salzberg. Identifying bacterial genes and endosymbiont DNA with Glimmer, Bioinformatics 23:6 (2007), 673-679.
[7] Hyatt, D., Chen, G. L., Locascio, P. F., Land, M. L., Larimer, F. W., & Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC bioinformatics, 11, 119. doi:10.1186/1471-2105-11-119
[8] Gene Prediction. Wikipedia. 2019. https://en.wikipedia.org/wiki/Gene_prediction
[9] Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990) "Basic local alignment search tool." J. Mol. Biol. 215:403-410. PubMed
[10] Benjamin Buchfink, Chao Xie & Daniel H. Huson, Fast and Sensitive Protein Alignment using DIAMOND, Nature Methods, 12, 59–60 (2015) doi:10.1038/nmeth.3176.
[11] HMMER: biosequence analysis using profile hidden Markov models. http://hmmer.org/
[12] Lowe, T. M., & Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids research, 25(5), 955–964. doi:10.1093/nar/25.5.955
[13] Laslett, D., & Canback, B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic acids research, 32(1), 11–16. doi:10.1093/nar/gkh152
[14] Transfer RNA. Wikipedia. (2020).
[15] Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., & Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic acids research, 35(9), 3100–3108. doi:10.1093/nar/gkm160
[16] Torsten Seemann. (2018). BAsic Rapid Ribosomal RNA Predictor. https://github.com/tseemann/barrnap
[17] Kate. M. (2017). How do mRNA, tRNA work together in translation to build protein? Socratic Q&A. https://socratic.org/questions/how-do-mrna-trna-and-rrna-work-together-in-translation-to-build-protein