Team II Gene Prediction Group
Team 2: Gene Prediction
Team Members: Danielle Temples, Kara Lee, Paarth Parekh, Shuting Lin
Introduction
Our Project
Purpose: Investigate an unknown outbreak pathogen using raw genome sequence data from the Centers for Disease Control and Prevention (CDC) foodborne illness surveillance outbreak investigations
Overall Objective: Identify and characterize the pathogenic organism, make recommendations for the outbreak control, and build a public webserver that automates the computational steps
Our Objective: From assembled genomes, predict genes or features using different prediction methods and evaluate selected tools on their accuracy and performance
What is Gene Prediction?
Identification of the regions of genomic DNA that encode genes, which are fragments of DNA that encodes a functional molecule:
- Protein-coding genes
- RNA genes
- May also include other functional elements (i.e. regulatory regions)
Prokaryotic Genome
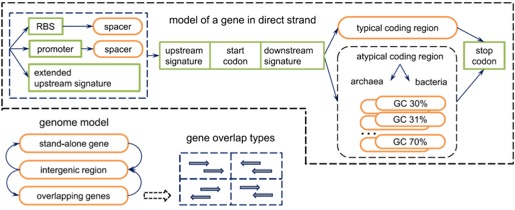
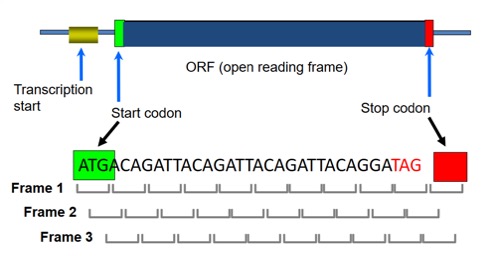
Prokaryotic Genomes have a high gene density and do not contain introns in their protein-coding regions. Genes are called Open Reading Frames or “ORFs” (include start & stop codon).
Prediction of prokaryotic genes tends to be relatively simpler with contiguous ORFs. However, overlapping ORFs and short genes can cause issues. Each gene is an ORF, but not every ORF is a gene.
Homology Methods
- Makes predictions via comparisons with sequences of previously known genes
- Based on extrinsic information
- Can be used to validate/support Ab Initio findings
- Limited by the use of no new knowledge
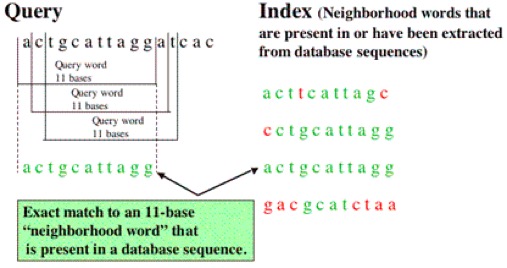
BLAST
Useful for dentifying species, locating domains, establishing phylogeny, DNA mapping, and comparison.
Algorithm:
- Break query into words of length W
- Align words with sequence in database & identify matches
- Calculate T score for matches
- Extend sequence in both directions until score falls below cutoff (HSPs)
- Report hits that meet or exceed BLAST cutoff for statistically significant hits
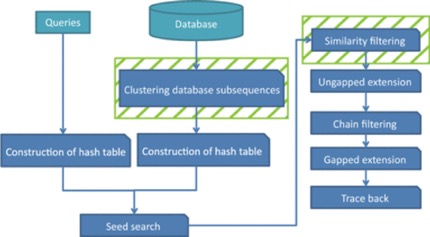
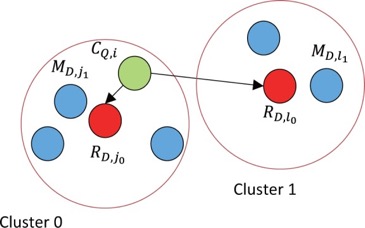
GHOSTZ
A new faster homology search method using database subsequence clustering.
Algorithm:
- Sequences are extracted from a database & similar ones are clustered
- Construct into hash tables
- Use hash tables to select seeds for the alignments from representative sequences in the clusters
- Distance between a query subsequence and cluster representative is calculated
- Lower bounds calculated
- Similarity Filtering – if computed lower bound is less than or equal to distance threshold, continue
Ab Initio Methods
- Inspect the input sequence and searches for traces of gene presence
- Simplest method is to inspect ORFs
- Relies on probability models & specific DNA motifs (signals)
- Markov Models and Dynamic Programming
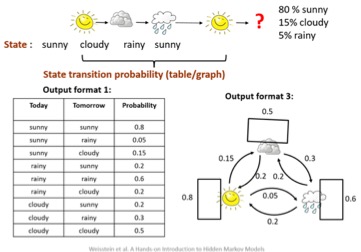
Hidden Markov Models
- Markov Model is a chain structured process where future states depend only on the present state
- Used to model randomly changing systems
- Hidden Markov Model (HMM) is a statistical Markov model with hidden states
- Viterbi Algorithm used to find the most likely sequence of hidden paths
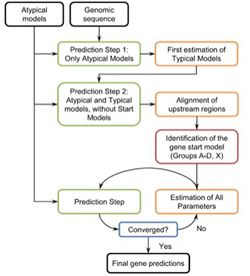
GeneMarkS-2
- Uses HMM and a self training algorithm (non supervised) to predict genes
- 5th Order HMM for coding and 2nd order for non-coding regions
- Identifies several different types of distinct sequence patterns
- The model which yields the highest log-odds score is selected
- Classifies the genome into 4 distinct groups:
- Group A: Typical Model of Prokaryotes having RBS sites having (SD)Consensus
- Group B: Atypical Model having RBS sites not having SD consensus
- Group C and D: Represent Bacterial and Archeal Genomes (Leaderless Transcription)
- Group X: Weak, Hard to classify regulatory signal patterns
- Stops after 10 iterations in the final prediction step, if it doesn’t converge
PRODIGAL
- Looks at GC bias for each of three codon positions and chooses the one with highest GC content
- Scores every start-stop pair above 90 bp in the entire genome based on simple GC codon statistics
- Penalizes or gives bonus to intergenic spaces according to gene distance
- Uses Dynamic Programming to force the program to choose between two heavily overlapping ORFS
- Sacrifices some genuine predictions to eliminate a much larger number of false identifications
Glimmer3
Tool Evaluation
Non-Coding RNA Methods
- Non-coding RNA (ncRNA) is an RNA molecule that is not translated into a protein
- Transfer RNAs (tRNAs), ribosomal RNAs (rRNAs) and small RNAs(sRNAs)
- Protein synthesis/Translation (tRNA and rRNA) & gene regulation (sRNA)
- Related to antibiotic resistance
ARAGORN
- Homology based tool
- Uses heuristic algorithms that score the tRNA and tmRNA genes based on their sequence and secondary structure similarities
- An effective tRNA search program, with sensitivity better than other current heuristic tRNA search algorithms
RNAmmer
- Ab Initio based tool
- Uses Hidden Markov Models trained on data from 5s rRNA database
- Fast with little loss of sensitivity, enabling the analysis of a complete genome in less than a minute
- Location of rRNAs can be predicted with a very high accuracy
Infernal
- Implementation of covariance models (CMs)
- RNA homology search based on accelerated profile HMM methods and HMM-banded CM alignment methods
- 100-fold faster RNA homology searches and ∼10,000-fold acceleration over exhaustive non-filtered CM searches