Team III Webserver Group
Team 3: Web Server
- File:Web Server - Team 3 Background and Strategy.pptx.pdf
- File:Web Server - Team 3 Final Presentation.pptx.pdf
Group Members - Aparna Maddala, Sonali Gupta, Ahish Sujay, Yiqiong Xiao, Allison Rozanski, Yuhua Zhang
Introduction
Problem Statement
- Makes our work as bioinformaticians accessible to a wider audience
- Is easy-to-use, requires little bioinformatics knowledge to get the analysis results
- Is visually informative, easy on eyes
Design Objectives
Users should be able to
- Go through the entire pipeline from genome assembly to comparative genomics
- Only execute individual steps
- Easily execute the remainder of the pipeline from any intermediate step
Architecture Design
Approaches
MVC Framework - MODEL
MySQL
- Corresponds to all the data-related logic that the user works with
- Constitutes the computation, execution and visualisations
- Completely abstracted from the user
- Flask does not support databases natively, gives flexibility on database choice best for application
- Flask-SQLAlchemy provides a flask friendly wrapper to the SQLAlchemy package.
- SQLAlchemy is an Object Relational Mapper (ORM) and supports several database engines including MySQL
- MySQL due to familiarity with server and installation
- Will Utilize MySQL for storing file paths and intermediate outputs
MVC Framework - View
Javascript, CSS, HTML
- Used for all the UI logic of the application
- Separates user from backend processing
MVC Framework - Controller
Python(Flask)
- Acts as an interface between Model and View components
- Responsible for validation of inputs from view and outputs from model before sending data to either of them
- Responsible for invocation of specific responses based on the requests received
- One of the most widely used Python based web frameworks
- Reasons for selection: Offers simple development; Easy to deploy; Fine-grained control; Provides flexible frameworks and is minimal, Familiarity with Flask
Web server
- Software that understands URLs (web addresses) and HTTP (the protocol your browser uses to view webpages).
- It can be accessed through the domain names of websites it stores, and delivers their content to the end-user's device.
Reverse Proxy
- Takes requests from the Internet and forwards them to servers in an internal network. Those making requests to the proxy may not be aware of the internal network.
Functionality
Pipeline
Genome Assembly
- Input files:Paired-end fastq files for Listeria monocytogenes
- Process:Perform quality control and trimming using fastp, Assemble genomes and plasmids using SPAdes
- Output files: HTML quality control report (Generated by MultiQC), FASTA contig files (Generated by SPAdes and Plasmid SPAdes)
Gene Prediction
- Process (Coding): Run both PRODIGAL and GeneMarkS-2; Use BLAST for validation and retrieve the coding output
- Process (Non-coding): Run ARAGORN, BARRNARP, and RNAmmer; Use Infernal for validation and retrieve the non-coding output
- Input: Assembled genomes and plasmids from the genome assembly pipeline
- Output files (Coding): FASTA files, GFF files
- Output files (Non-coding): FASTA files, GFF files
Functional Annotation
- Input: 50 files from gene prediction in .fna format
- Processes: Cluster using UCLUST; eggNOG, CARD ,VFDB, PilerCR, SignalP and HMMTOP; merge functional annotations
- Output: 50 files in .gff and summary of annotation results i.e. annotation; count for each tool, .gff display and significant antibiotic and resistance genes present
Comparative Genomics
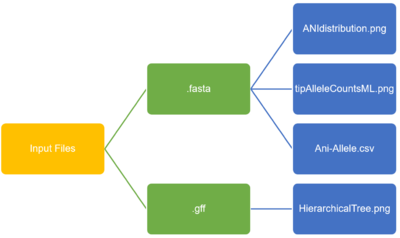
- Comparative genomics is an essential step in food borne outbreak analysis. People usually use different level of bioinformatics tools to find the distance of isolates. From the study of our comparative genomics group, we found that the results from Average Nucleotide Identity (ANI), allele phylogenetic tree, and annotated hierarchical tree provide the most information.
- The comparative genomics webserver can take either assembled fasta file from genome assembly analysis or gff file from functional annotation analysis, 10 files maximum. The program will detect the type of input automatically. And the outputs are ANI distribution figure and maximum likelihood allele phylogenetic tree when input are fasta files; the output is a hierarchical tree figure when inputs are gff files.
Webserver Walk through
Please find our web server here: http://predict2020t3.biosci.gatech.edu/
We used SQLite3 database and integrated it with the flask app using SQLAlchemy. The DB table was designed to have the following fields :
- 1. Job ID (primary key)
- 2. Pipeline execution completed
- 3. Email sent
Job ID is a unique random identifier for each request submitted.
The email sender runs in an infinite loop and scans the database table to identify the Job IDs which completed the pipeline execution but the email haven't been sent. It then send a download link for those job IDs.
References
https://compgenomics2020.biosci.gatech.edu/Team_III_Comparative_Genomics_Group