Team I Functional Annotation Group
Team 1 Functional Annotation
Team members: Maria Ahmad, Shuheng Gan, Manasa Vegesna, Hyeonjeong Cheon, and Kenji Gerhardt
Introduction
Background
Functional annotation is the process of associating predicted genes with their functional role in a cell. This includes the type of gene (translated/untranslated), their location within the cell, and their chemical and biological roles. These can be derived both by comparison to similar, already annotated genes with known (or anticipated) functions, or through ab initio annotation that relies on existing models, but which does not rely on databases of other annotated genes.
Objective
Our goal in this step is to functionally annotate the genes supplied to us by the gene prediction group.
Data
The genomes supplied to us were identified during assembly as E. coli. E. coli is the most highly studied microorganism: at present, NCBI contains 1072 complete E. coli genomes in its genome database, along with thousands of additional chromosomes, contigs, and scaffolds. This depth of study extends to its genes, where multiple pathogenic strains are identified and characterized well.
Pipeline
Pipeline image will be inserted
Clustering
USearch
Methods
Ab-initio Approach
Ab-Initio Tools predict and annotate different regions of the prokaryotic genome using 1) sequence composition, 2) likelihoods within the gene models, 3) gene content, and 4) ignal Detection. Ab-Initio Approach can be used for finding new genes, and no external data or evidence is needed for the prediction. However, it is limited by the presence of False Positives in the predicted data as well as over-prediction of small genes.
PSORTb
remove if not needed
SignalP
remove if not needed
LipoP
remove if not needed
TatP
remove if not needed
TMHMM
remove if not needed
Phobius
remove if not needed
CRISPRCasFinder
remove if not needed
PILER-CR
remove if not needed
CRT
remove if not needed
Homology-based Tools
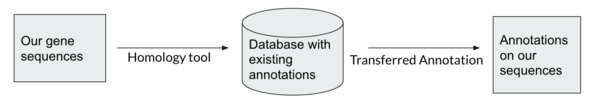
Homology between genes means they share ancestry. Homologous genes that have recently diverged usually share function. We are looking to transfer annotation on known genes to our predicted genes by finding homology genes. Gene databases are collections of annotated genes. When we search a gene against a database, the search is looking for homology between our gene sequences and those in the database to determine what our genes’ function will be.
Homology-based tools are more accurate and reliable than Ab-initio tools, and can be targeted for specific purposes, e.g. antibiotic resistance genes. However, homology tools are dependent on existing annotations and on what databases are being searched.
Prokka
- Prokka is a command line software tool for the rapid annotation of prokaryotic genomes in about 10 min on a desktop computer.
- Features include coding sequences (Prodigal), rRNA (RNAmmer), tRNA (Aragorn), signal peptides (SignalP), and non-coding RNA (Infernal) and including these options are flexible.
- The typical input file is FASTA format and output contains 12 output files: .err, .fsa, .sqn, .val, .faa, .gbf, .tbl, .ffn, .gff, .tsv, .fna, .log, .txt
Command:
prokka <input_file> --outdir <output directory> --prefix <file prefixes> --kingdom Bacteria --locustag --norrna --notrna
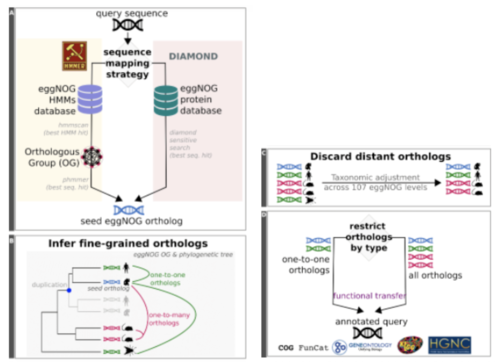
EggNOG-mapper
- Performes fast functional annotation of genes and proteins
- Uses precomputed orthologous groups and phylogenies from the eggNOG database which consists of Orthologous Groups( OGs) of proteins at taxonomic levels.
- Has a mode recommended for large datasets called DIAMOND. Diamond mode searches for the best seed ortholog located in the eggNOG protein database, and is faster than the other mode HMM. The HMM mode is comprised of a collection of precompiled hidden Markov models.
To download bacteria databases:
python2.7 download_eggnog_data.py bact
Command:
python emapper.py -i <input_file> --output <output_file> -m diamond -d bact -o <output directory>
InterProScan
- InterProScan is function annotation tool that scans sequences against InterPro protein signature databases. It classifies proteins into families and predict domains and important sites.
- Signatures are predictive models constructed from multiple sequence alignments that can be used to classify proteins. Using protein signatures is often a more sensitive way of identifying protein function than pairwise sequence similarity searches, such as BLAST.
- InterPro combines four types of protein signatures (patterns, profiles, fingerprints and HMMs) from multiple databases.
Download (https://github.com/ebi-pf-team/interproscan/wiki/HowToDownload)
- download interproscan and most database
wget ftp://ftp.ebi.ac.uk/pub/software/unix/iprscan/5/5.42-78.0/interproscan-5.42-78.0-64-bit.tar.gz
wget ftp://ftp.ebi.ac.uk/pub/software/unix/iprscan/5/5.42-78.0/interproscan-5.42-78.0-64-bit.tar.gz.md5
md5sum -c interproscan-5.42-78.0-64-bit.tar.gz.md5
- download Panther
cd [InterProScan5 home]/data/
wget ftp://ftp.ebi.ac.uk/pub/software/unix/iprscan/5/data/panther-data-14.1.tar.gz
wget ftp://ftp.ebi.ac.uk/pub/software/unix/iprscan/5/data/panther-data-14.1.tar.gz.md5
md5sum -c panther-data-14.1.tar.gz.md5
Requirement (https://github.com/ebi-pf-team/interproscan/wiki/InstallationRequirements)
- - Perl 5.22
- - Python 3
- - Java 11
Command
/interproscan.sh -i <input_file_name> -dp -d <output_directory>
default: run all 14 databases default outputs: input_file_name.gff3 input_file_name.tsv input_file_name.xml
DeepARG
Results
Reference
[1] Joshi, T., & Xu, D. (2007). Quantitative assessment of relationship between sequence similarity and function similarity. BMC genomics, 8(1), 222.
[2] Zou, Q., Lin, G., Jiang, X., Liu, X., & Zeng, X. (2020). Sequence clustering in bioinformatics: an empirical study. Briefings in bioinformatics, 21(1), 1-10.
[3] Nielsen, H., Tsirigos, K. D., Brunak, S., & von Heijne, G. (2019). A brief history of protein sorting prediction. The protein journal, 38(3), 200-216.
[4] Viklund, H., & Elofsson, A. (2004). Best α‐helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Science, 13(7), 1908-1917.
[5] Romine, M. F. (2011). Genome-wide protein localization prediction strategies for gram negative bacteria. BMC genomics, 12(S1), S1.
[6] Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., ... & Brinkman, F. S. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics, 26(13), 1608-1615.
[7] Armenteros, J. J. A., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., ... & Nielsen, H. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature biotechnology, 37(4), 420-423.
[8] Juncker, A. S., Willenbrock, H., Von Heijne, G., Brunak, S., Nielsen, H., & Krogh, A. (2003). Prediction of lipoprotein signal peptides in Gram‐negative bacteria. Protein Science, 12(8), 1652-1662.
[9] Bendtsen, J. D., Nielsen, H., Widdick, D., Palmer, T., & Brunak, S. (2005). Prediction of twin-arginine signal peptides. BMC bioinformatics, 6(1), 167.
[10] Käll, L., Krogh, A., & Sonnhammer, E. L. (2004). A combined transmembrane topology and signal peptide prediction method. Journal of molecular biology, 338(5), 1027-1036.
[11] Krogh, A., Larsson, B., Von Heijne, G., & Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology, 305(3), 567-580.
[12] Couvin, D., Bernheim, A., Toffano-Nioche, C., Touchon, M., Michalik, J., Néron, B., ... & Pourcel, C. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic acids research, 46(W1), W246-W251.
[13] Mao, F., Dam, P., Chou, J., Olman, V., & Xu, Y. (2009). DOOR: a database for prokaryotic operons. Nucleic acids research, 37(Database issue), D459–D463. https://doi.org/10.1093/nar/gkn757
[14] Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., & Jin, Q. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic acids research, 33(Database issue), D325–D328. https://doi.org/10.1093/nar/gki008
[15] eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Jaime Huerta-Cepas, Damian Szklarczyk, Kristoffer Forslund, Helen Cook, Davide Heller, Mathias C. Walter, Thomas Rattei, Daniel R. Mende, Shinichi Sunagawa, Michael Kuhn, Lars Juhl Jensen, Christian von Mering, and Peer Bork.Nucl. Acids Res. (04 January 2016) 44 (D1): D286-D293. doi: 10.1093/nar/gkv1248
[16] McArthur et al. 2013. The Comprehensive Antibiotic Resistance Database. Antimicrobial Agents and Chemotherapy, 57, 3348-57.
[17] Nucleic Acids Res. 2009 Jan;37(Database issue):D443-7. doi: 10.1093/nar/gkn656. Epub 2008 Oct 2.
[18] Arndt, D., Grant, J., Marcu, A., Sajed, T., Pon, A., Liang, Y., Wishart, D.S. (2016) PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res., 2016 May 3.
[19] Arango-Argoty, G., Garner, E., Pruden, A. et al. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 6, 23 (2018). https://doi.org/10.1186/s40168-018-0401-z
[20] Ashburner et al. Gene ontology: tool for the unification of biology. Nat Genet. May 2000;25(1):25-9.
[21] Prediction of lipoprotein signal peptides in Gram-negative bacteria.A. S. Juncker, H. Willenbrock, G. von Heijne, H. Nielsen, S. Brunak and A. Krogh.Protein Sci. 12(8):1652-62, 2003
[22] Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., Von Mering, C., & Bork, P. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Molecular biology and evolution, 34(8), 2115-2122.
[23] Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics, 30(14), 2068-2069.
[24] Arango-Argoty, G., Garner, E., Pruden, A., Heath, L. S., Vikesland, P., & Zhang, L. (2018). DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome, 6(1), 1-15.
[25] Finn, R. D., Attwood, T. K., Babbitt, P. C., Bateman, A., Bork, P., Bridge, A. J., ... & Gough, J. (2017). InterPro in 2017—beyond protein family and domain annotations. Nucleic acids research, 45(D1), D190-D199.
[26] Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., & Madden, T. L. (2009). BLAST+
[27] Buchfink, B., Xie, C., & Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nature methods, 12(1), 59.