Team II Comparative Genomics Group: Difference between revisions
| Line 93: | Line 93: | ||
Tools to be tested: | Tools to be tested: | ||
*kSNP3.0 (2) | *kSNP3.0 (2) | ||
** | ** Optimal for situations where whole genome alignments don't work | ||
** k-mer-based approaches are alignment-free and have a faster runtime | |||
** Multiple kSNP versions have been created and thoroughly tested | |||
*Lyve-SET (3) | *Lyve-SET (3) | ||
** MSA-based approach (computationally expensive) | |||
** Consistent performance according to literature | |||
** Has a higher sensitivity, specificity, and average Sn and Sp than kSNP | |||
=='''Outbreak Analysis Results'''== | =='''Outbreak Analysis Results'''== | ||
Revision as of 09:09, 28 March 2020
Team 2: Comparative Genomics
Team Members: Kara Keun Lee, Courtney Astore, Kristine Lacek, Ujani Hazra, Jayson Chao
Class Presentations
Introduction
What is Comparative Genomics?
Once genomes are fully assembled and annotated, outbreak analysis can begin via comparative genomics. Generally, metadata ascertained from gene prediction and annotation can be used to map the relatedness of multiple isolates. Combined with epidemiological data, a given outbreak can be mapped back to a particular source (patient zero) and tracked to determine which strains are outbreak isolates and which are sporadic cases. Furthermore, phenotypic features such as virulence, antibiotic resistance, and pathogenicity can be determined. Compilation of these data allows for recommendations to be made on behalf of human impact, treatment strategy, and management methods to address further spread.
Our Data
Our genomic data comes from 50 isolates of C. jejuni from an outbreak of foodborne illnesses. The genomes are assembled and fully annotated.
Our epidemiological data include times, locations, and ingested foods of each case.
Pipeline Overview
Objectives
- Identify kinds of strains (outbreak vs. sporadic)
- Construct phylogeny demonstrating which isolates are related and which differ
- Determine the source of the outbreak
- Map virulence and antibiotic resistance features of outbreak isolates
- Compile recommendations for outbreak response and treatment
Overview of Techniques
When performing phylogenomics, there are many options by which one can classify similarities and differences across the genome. Our approach utilizes tools from three different techniques.
ANI
MLST
MLST or Multi-locus Sequence Typing identifies a set of loci (housekeeping genes) in the genome and compares each locus in a genome against the set of loci. It estimates the relationships between bacteria based on allelic variations in specific loci than their nucleotide sequences. MLST data can be used to investigate evolutionary relationships among bacteria. However, the sequence conservation of the housekeeping genes limits the discriminatory power of MLST in differentiating bacterial strains.
There are several types of MLST:
Whole-genome MLST (wgMLST): All loci of a given isolate compared to equivalent loci in other isolates (typing scheme based on a few thousand genes).
- Creates wgMLST tree (different styles exist)
- Minimum spanning tree = circles with sizes indicative of the frequency of ST and distance showed on connecting lines
Core-genome MLST (cgMLST): Focused on only the core elements of the genomes of a group of bacteria (typing scheme based on a few hundred genes).
7-Gene MLST: Chooses 7 loci in the genome and compare all genomes to these 7 loci.
- Profile of alleles (“sequence type” or ST) by calling the alleles
- Genome assembly optional – there are assembly free methods
- Creates a phylogeny
Ribosomal MLST (rMLST): Based on 53 loci that code for ribosomal proteins present in most bacteria.
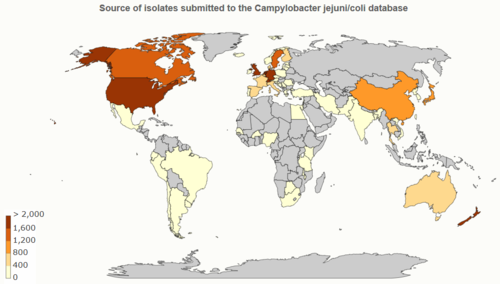
Database: PubMLST
PubMLST (Public databases for molecular typing and microbial genome diversity) for Campylobacter jejuni/coli (as of 17MAR2020):
- 98,017 isolates
- 50,138 genomes
- 1,286,733 alleles
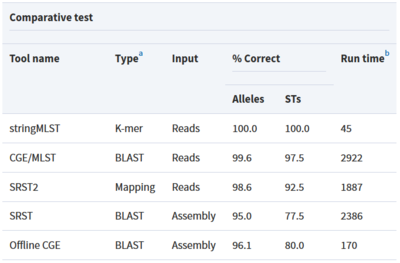
Tool: stringMLST
stringMLST is a tool for detecting the MLST of an isolate directly from the genome sequencing reads.
- Predicts the ST of an isolate in a complete assembly and alignment-free manner
- Downloads and builds databases from pubMLST using the most recent allele and profile definitions
- Faster algorithm compared to traditional MLST tools that maintain high accuracy
Tool: MentaLiST
A k-mer based MLST caller designed specifically for handling large MLST schemes.
- Capable of dealing with MLST schemes with up to thousands of genes while requiring limited computational resources
- MLST calling that does not require pre-assembled genomes, working directly with the raw WGS data, and also avoids costly pre-processing steps (i.e. contig assembly or read mapping onto a reference)
- Follows the general principle of k-mer counting, introduced in stringMLST, with some data compression improvements that lead to much smaller database sizes and a faster running time
Tool: ARIBA
Assembly based tool, primarily developed for identifying Anti-Microbial Resistance - associated genes and single nucleotide polymorphisms directly from short reads
- Provides inbuilt support for and functionality for multi-locus sequence typing (MLST) using data from PubMLST
- Provides inbuilt support for PlasmidFinder and VFDB (Virulence Factor Databases)
- Can be used in the study of Virulence Profile and AMR features along with the results from the Functional Annotation group
SNP-based
SNP stands for Single Nucleotide Polymorphism, meaning that certain alleles have two or three possibilities as to which base is at a given locus. As SNPs accumulate through de novo mutations and are passed down through generations, comparing a given isolate's SNPs to other isolates and a reference genome allow ascertainment of phylogenetic distance between samples(1). Tools have been developed to compare bases position by position (SNP-calling) and create matrices to compute relatedness between samples based on common SNPs.
Generalized Algorithm Overview:
- Pre-processing and read cleaning
- Mapping
- SNP calling against a reference genome
- Phylogeny generation based on SNP profiles
Tools to be tested:
- kSNP3.0 (2)
- Optimal for situations where whole genome alignments don't work
- k-mer-based approaches are alignment-free and have a faster runtime
- Multiple kSNP versions have been created and thoroughly tested
- Lyve-SET (3)
- MSA-based approach (computationally expensive)
- Consistent performance according to literature
- Has a higher sensitivity, specificity, and average Sn and Sp than kSNP
Outbreak Analysis Results
Source of Outbreak
Human Impact
Treatment Strategy
CDC Recommendations
Works Cited
1.https://cba.anu.edu.au/news-events/snps-population-and-phylo-genomics
2.https://academic.oup.com/bioinformatics/article/31/17/2877/183216
3.https://github.com/lskatz/lyve-SET.Katz et al. (2017) A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology for foodborne pathogens. Frontiers in Microbiology 8: 375.