Team II Functional Annotation Group: Difference between revisions
| (83 intermediate revisions by 4 users not shown) | |||
| Line 21: | Line 21: | ||
== '''Clustering'''== | == '''Clustering'''== | ||
* Significant sequence similarity implies shared ancestry that often leads to shared function | |||
* Clustering such sequences can reduce repeat queries in homology-based annotations | |||
* Reducing repeats improves speed and storage costs | |||
[[File:cluster.png|border|400px|Clustering]] | |||
===[http://weizhongli-lab.org/cd-hit/ '''CD-HIT''']=== | |||
* Widely used program for clustering and comparing protein or nucleotide sequences | |||
* Very fast and can handle extremely large databases | |||
./cd-hit -i [input file] -o [output file name] | |||
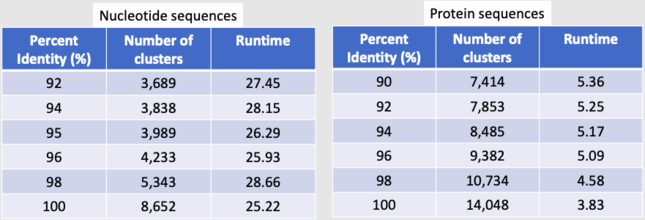
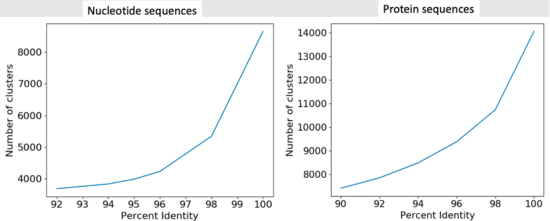
* We were able to get 90305 protein sequences down to 7414 representative clusters. | |||
* We were able to get 90305 DNA sequences down to 3989 representative clusters. | |||
== '''Homology Methods'''== | == '''Homology Methods'''== | ||
| Line 43: | Line 56: | ||
* Identifying these may enhance our knowledge of gene regulation & function which is a key addition to genome annotation | * Identifying these may enhance our knowledge of gene regulation & function which is a key addition to genome annotation | ||
===[https://github.com/ | ===[https://github.com/eggnogdb/eggnog-mapper '''eggNog-Mapper''']=== | ||
* ''' | * '''E'''volutionary '''G'''enealogy of '''G'''enes: '''N'''on-supervised '''O'''rthologous '''G'''roups | ||
* | * Functional Annotation of large sequence sets by fast orthology assignments using precomputed clusters and phylogenies from the eggNOG database | ||
* | * Use HMM-based search and Diamond based searches to lead to the best seed ortholog in eggNOG | ||
[[File: | * Orthology predictions for functional annotation results in a higher precision than traditional homology searches by avoiding transferring annotations from close paralogs | ||
[[File:eggy.jpg|border|550px|eggNog-mapper Algorithm]] | |||
Download the necessary databases. This will fetch and decompress all precomputed eggNOG data into the data/ directory. | |||
python2 ./download_eggnog_data.py | |||
Run eggNog-mapper for clustering results against eggNOG bacteria database. | |||
python2 ./emapper.py -i <cluster> --output <result> -d bact -m diamond | |||
==='''VFDB'''=== | ===[http://www.mgc.ac.cn/VFs/main.htm '''VFDB''']=== | ||
* '''V'''irulence '''F'''actor '''D'''ata'''B'''ase | * '''V'''irulence '''F'''actor '''D'''ata'''B'''ase | ||
* Provide virulence structure features, functions, and mechanisms used to allow pathogens to conquer new niches and circumvent host defense mechanisms | * Provide virulence structure features, functions, and mechanisms used to allow pathogens to conquer new niches and circumvent host defense mechanisms | ||
* BLAST based identification of virulence genes | * BLAST based identification of virulence genes | ||
Download VFDB '''core dataset''' (includes genes associated with experimentally verified VFs only) and convert into BLAST database. | |||
makeblastdb -in VFDB_db -dbtype 'nucl' -out <db_name> | |||
Run BLASTn for clustering results against VFDB database. | |||
blastn -db <db_name> -query <cluster> -out <result> -max_hsps 1 -max_target_seqs 1 -outfmt "6 qseqid length qstart qend sstart send evalue bitscore stitle" -perc_identity 100 -num_threads 5 | |||
==='''CARD'''=== | ===[https://card.mcmaster.ca/ '''CARD''']=== | ||
* '''C'''omprehensive '''A'''ntibiotic '''R'''esistance '''D'''atabase | * '''C'''omprehensive '''A'''ntibiotic '''R'''esistance '''D'''atabase | ||
* Provides data, models, and algorithms relating to the molecular basis of antimicrobial resistance | * Provides data, models, and algorithms relating to the molecular basis of antimicrobial resistance | ||
* Can be used for the analysis of genome sequences using the Resistance Gene Identifier | * Can be used for the analysis of genome sequences using the Resistance Gene Identifier | ||
== '''''Ab Initio'' | Install the RGI software using conda & download CARD database: | ||
wget https://card.mcmaster.ca/latest/data | |||
tar -xvf data ./card.json | |||
Build a local database: | |||
rgi load --card_json <path to card.json> --local | |||
Run RGI main with protein sequences: | |||
rgi main -i <path to cluster.faa> -o <output_file_name> -t protein --local | |||
Create a tab-delimited file from rgi results: | |||
rgi tab -i <path to output_file_name.json> | |||
===[http://www.microbesonline.org/operons/ '''MicrobesOnline''']=== | |||
Predicts whether pairs of adjacent genes that are on the same strand are in the same operon, based on the intergenic distance between them, whether orthologs of the genes are near each other in other genomes, and their predicted functions. | |||
* Download all the operon tables for Campylobacter jejuni. | |||
* Use GID to retrieve the protein reference sequences. | |||
* Create a database using BLAST: | |||
makeblastdb -in <fasta file > -dbtype prot -out <database> | |||
* Query the clustered sequences with the reference protein database: | |||
blastp -query cdhit/faa_rep_seq.faa -db tmp/db_operon -evalue 0.01 -max_target_seqs 1 -max_hsps 1 -outfmt 6 -out tmp/hits_0.01.txt -num_threads 5 | |||
== '''''Ab Initio'' Methods'''== | |||
==='''Categories'''=== | |||
<u>Transmembrane Proteins (Cell Membrane and Outer Membrane):</u> | |||
* Bacteria have the ability to export effector proteins in membranes of eukaryotic host | |||
* Integral membrane protein that function as gates or docking sites that allow or prevent the entry or exit of materials across the cell membrane | |||
== ''' | <u>Signal Peptides:</u> | ||
* Guide secretory proteins to find their correct locations outside the cell membrane for signal transduction | |||
<u>CRISPR:</u> | |||
* Provides immunity to the bacteria against Bacteriophages | |||
* Contributes to the Virulence and Pathogenicity of the bacteria | |||
===[http://www.cbs.dtu.dk/services/TMHMM/ '''TMHMM2''']=== | |||
* Uses Hidden Markov Model approach | |||
* Outputs the number of transmembrane helices predicted, the position of each residue with respect to the cell membrane (outside/inside the cell, transmembrane segment), and optional graphical output visualizing the predicted helix | |||
Input a multifasta file into TMHMM. | |||
tmhmm <input multifasta file> > <output file> | |||
===[https://www.drive5.com/pilercr/ '''PilerCR''']=== | |||
* Rapid identification and classification of CRISPR repeats | |||
* High sensitivity and high specificity | |||
[[File:pilercr.png|border|Structure of a CRISPR Array]] | |||
Input a multifasta file into PileCR. | |||
pilercr –in <input multifasta file> -out <output file> -noinfo –quiet | |||
===[http://www.cbs.dtu.dk/services/SignalP/ '''SignalP''']=== | |||
* Uses a deep neural network algorithm | |||
* Predicts presence of signal peptides and location of cleavage sites | |||
* Does not determine lipoproteins | |||
[[File:signalp.jpg|border|400px|SignalP Architecture]] | |||
Input a fasta file into SignalP. | |||
signalp –fasta <input_sequence_file> -org gram- -format short –gff3 | |||
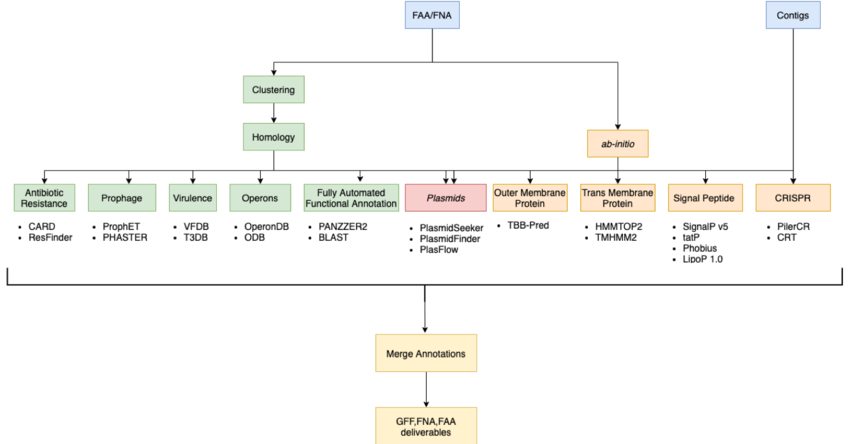
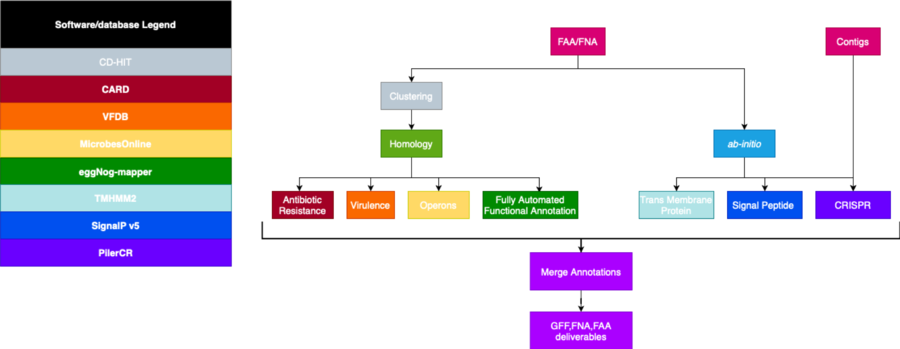
== '''Initial Pipeline'''== | |||
[[File:initial_pipe.png|border|850px|Initial Pipeline]] | |||
== '''Results'''== | == '''Results'''== | ||
== ''' | |||
== ''' | ==='''Clustering'''=== | ||
[[File:clust1.png|border|645px|Clustering Results 1]] [[File:clust2.png|border|550px|Clustering Results 2]] | |||
==='''''Ab Initio'''''=== | |||
<u>Transmembrane Proteins</u> | |||
* Minimum = 810 | |||
* Maximum = 1,049 | |||
* Average = 928.8 | |||
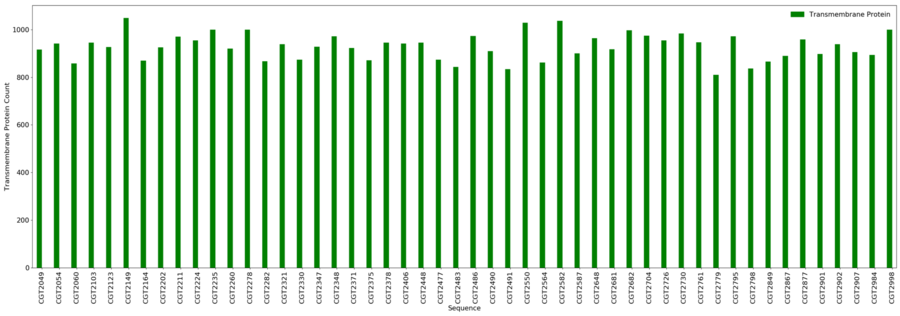
[[File:transmem.png|border|900px|Transmembrane Protein Results]] | |||
<u>Signal Peptides</u> | |||
* Minimum = 71 | |||
* Maximum = 99 | |||
* Average = 94.3 | |||
<u>Lipoproteins</u> | |||
* Minimum = 43 | |||
* Maximum = 67 | |||
* Average = 52.8 | |||
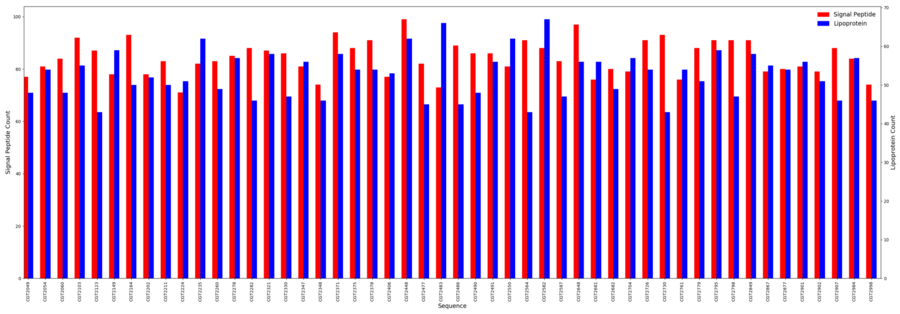
[[File:lipoprot.png|border|900px|Signal Peptide & Lipoprotein Results]] | |||
<u>CRISPR</u> | |||
* Minimum = 0 | |||
* Maximum = 2 | |||
* Average = 0.8 | |||
[[File:crisprres.png|border|900px|PilerCR Results]] | |||
==='''Homology'''=== | |||
<u>Antibiotic Resistance</u> | |||
* Minimum = 1 | |||
* Maximum = 10 | |||
* Average = 3.8 | |||
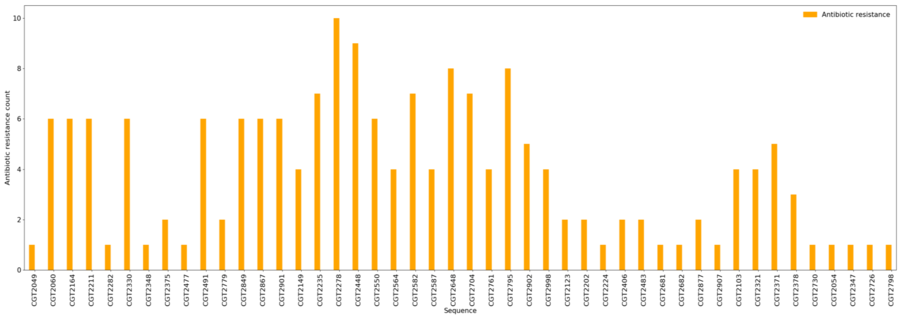
[[File:card.png|border|900px|CARD Results]] | |||
<u>VFDB Ouput</u> | |||
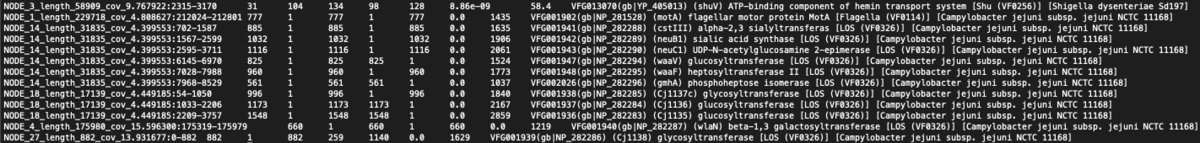
[[File:vfdboutput.png|border|1200px|VFDB Output]] | |||
<u>Virulence Factors</u> | |||
* Total = 132 | |||
* Minimum = 1 | |||
* Maximum = 13 | |||
* Average = 3.1 | |||
[[File:vfax.png|border|900px|VFDB Results]] | |||
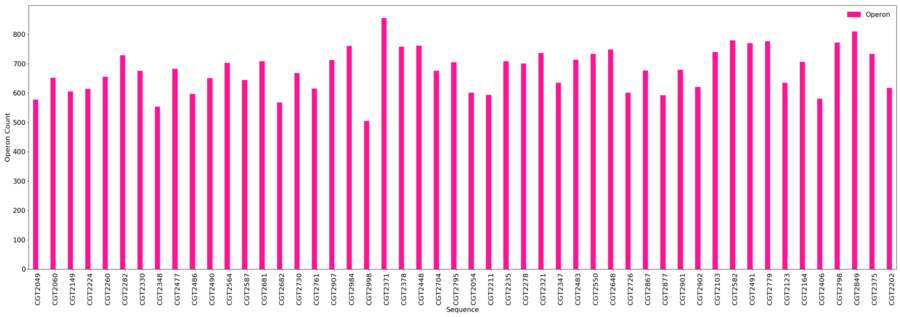
<u>Operons</u> | |||
* Minimum = 505 | |||
* Maximum = 856 | |||
* Average = 678.1 | |||
[[File:operons.png|border|900px|Microbes Online Results]] | |||
<u>eggNog-mapper Ouput</u> | |||
EggNog-mapper provides useful functional annotations such as seed eggNOG ortholog, ''seed ortholog evalue/score, predicted taxonomic group, predicted protein name, GO terms, KEGG ko, KEGG Pathways, eggnog OG, COG Functional Category, free text descriptions'', and many more. The descriptions of the '''COG Functional Categories''' are shown below. | |||
[[File:eggout.png|border|900px|eggNog Output]] [[File:COG.jpg|border|270px|COG Functional Categories]] | |||
<u>eggNog-mapper Results</u> | |||
* Total = 44,098 | |||
* Minimum = 669 | |||
* Maximum = 1087 | |||
* Average = 881.9 | |||
[[File:eggres.png|border|900px|eggNog Results]] | |||
== '''Final Pipeline'''== | |||
[[File:completoo.png|border|900px|Final Pipeline]] | |||
== '''References'''== | == '''References'''== | ||
Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1899501/ | |||
Genomic Characterization of Campylobacter jejuni Strain M1 . https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2928727/ | |||
A Genome-Wide Association Study to Identify Diagnostic Markers for Human Pathogenic Campylobacter jejuni Strains https://www.frontiersin.org/articles/10.3389/fmicb.2017.01224/full | |||
Campylobacter jejuni transcriptional and genetic adaptation during human infection https://centerforimmunizationresearch.org/wp-content/uploads/2018/08/Campylobacter-jejuni-transcriptional-and-genetic-adaptation-during-human-infection.pdf | |||
A proteome-wide protein interaction map for Campylobacter jejuni https://link.springer.com/article/10.1186/gb-2007-8-7-r130 | |||
Li, Weizhong & Fu, Limin & Wu, Sitao & Wooley, John. (2012). Ultrafast clustering algorithms for metagenomic sequence analysis. Briefings in bioinformatics. 13. 10.1093/bib/bbs035. | |||
Xavier, Basil Britto et al. “Consolidating and Exploring Antibiotic Resistance Gene Data Resources.” Journal of clinical microbiology vol. 54,4 (2016): 851-9. doi:10.1128/JCM.02717-15 | |||
Huerta-Cepas, Jaime & Forslund, Sofia & Coelho, Luis Pedro & Szklarczyk, Damian & Jensen, Lars & von Mering, Christian & Bork, Peer. (2017). Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Molecular Biology and Evolution. 34. 10.1093/molbev/msx148. | |||
Alcock, Brian P et al. "CARD 2020: Antibiotic Resistome Surveillance With The Comprehensive Antibiotic Resistance Database". Nucleic Acids Research, 2019. Oxford University Press (OUP), doi:10.1093/nar/gkz935 | |||
M. N. Price, E. J. Alm, and A. P. Arkin (2005). Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication.. Nucleic Acids Research 33:3224-3234. | |||
Marasini, Daya et al. “Phylogenetic Relatedness Among Plasmids Harbored by Campylobacter jejuni and Campylobacter coli Isolated From Retail Meats.” Frontiers in microbiology vol. 9 2167. 12 Sep. 2018, doi:10.3389/fmicb.2018.02167 | |||
Carattoli, Alessandra et al. “In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing.” Antimicrobial agents and chemotherapy vol. 58,7 (2014): 3895-903. doi:10.1128/AAC.02412-14 | |||
Marasini, Daya et al. “Phylogenetic Relatedness Among Plasmids Harbored by Campylobacter jejuni and Campylobacter coli Isolated From Retail Meats.” Frontiers in microbiology vol. 9 2167. 12 Sep. 2018, doi:10.3389/fmicb.2018.02167 | |||
Carattoli, Alessandra et al. “In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing.” Antimicrobial agents and chemotherapy vol. 58,7 (2014): 3895-903. doi:10.1128/AAC.02412-14 | |||
Roosaare M, Puustusmaa M, Möls M, Vaher M, Remm M. 2018. PlasmidSeeker: identification of known plasmids from bacterial whole genome sequencing reads. PeerJ6:e4588 https://doi.org/10.7717/peerj.4588 | |||
Weizhong Li, Lukasz Jaroszewski, Adam Godzik, Clustering of highly homologous sequences to reduce the size of large protein databases , Bioinformatics, Volume 17, Issue 3, March 2001, Pages 282–283, https://doi.org/10.1093/bioinformatics/17.3.282 | |||
Steinegger, M., Söding, J. Clustering huge protein sequence sets in linear time. Nat Commun 9, 2542 (2018). https://doi.org/10.1038/s41467-018-04964-5 | |||
Benjamin T James, Brian B Luczak, Hani Z Girgis, MeShClust: an intelligent tool for clustering DNA sequences, Nucleic Acids Research, Volume 46, Issue 14, 21 August 2018, Page e83, https://doi.org/10.1093/nar/gky315 | |||
Robert C. Edgar, Search and clustering orders of magnitude faster than BLAST, Bioinformatics, Volume 26, Issue 19, 1 October 2010, Pages 2460–2461, https://doi.org/10.1093/bioinformatics/btq461 | |||
Soto SM. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence. 2013;4(3):223–229. doi:10.4161/viru.23724 | |||
Latest revision as of 08:42, 24 April 2020
Team 2: Functional Annotation
Team Members: Danielle Temples, Courtney Astore, Rhiya Sharma, Ujani Hazra, Sooyoun Oh
Introduction
What is Functional Annotation?
The practice of putting biological meaning to coding genes (genes that encode proteins) and their corresponding protein sequences. Such annotations can be derived using homology and ab initio based approaches, which will be further explained in subsequent sections.
Objective: Perform a full functional annotation on the genes and proteins determined by the Gene Prediction group that is relevant to C. jejuni
Homology Approaches
- Determine function via sequence similarity to already functionally annotated sequences
- Limited by what we already know.
Ab Initio Approaches
- Determine function via predictive model without comparing to existing sequences
- Based on laws of nature
- Difficult to verify without experiments
Data Overview
We received 50 fna and 50 faa files from the gene prediction group. The 50 fna files are multifasta files representing each genome. The 50 faa files are multifasta files representing each proteome.
Clustering
- Significant sequence similarity implies shared ancestry that often leads to shared function
- Clustering such sequences can reduce repeat queries in homology-based annotations
- Reducing repeats improves speed and storage costs
CD-HIT
- Widely used program for clustering and comparing protein or nucleotide sequences
- Very fast and can handle extremely large databases
./cd-hit -i [input file] -o [output file name]
- We were able to get 90305 protein sequences down to 7414 representative clusters.
- We were able to get 90305 DNA sequences down to 3989 representative clusters.
Homology Methods
Categories
Prophage:
- Play an important role in the evolution of bacterial genomes and their pathogenicity
- Can change or knock out gene functions; alter gene expression
Virulence:
- A pathogen's ability to infect or damage a host
- Ex: toxins, surface coats that inhibit phagocytosis, surface receptors that bind to host cells
Fully Automated Functional Annotation:
- Tools that annotate a spectrum of features related to the function
Antibiotic Resistance
- When bacteria develop the ability to defeat the drugs designed to kill them
- Leads to higher medical costs, prolonged hospital stays, and increased mortality
Operons:
- A functional unit of transcription and genetic regulation
- Identifying these may enhance our knowledge of gene regulation & function which is a key addition to genome annotation
eggNog-Mapper
- Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups
- Functional Annotation of large sequence sets by fast orthology assignments using precomputed clusters and phylogenies from the eggNOG database
- Use HMM-based search and Diamond based searches to lead to the best seed ortholog in eggNOG
- Orthology predictions for functional annotation results in a higher precision than traditional homology searches by avoiding transferring annotations from close paralogs
Download the necessary databases. This will fetch and decompress all precomputed eggNOG data into the data/ directory.
python2 ./download_eggnog_data.py
Run eggNog-mapper for clustering results against eggNOG bacteria database.
python2 ./emapper.py -i <cluster> --output <result> -d bact -m diamond
VFDB
- Virulence Factor DataBase
- Provide virulence structure features, functions, and mechanisms used to allow pathogens to conquer new niches and circumvent host defense mechanisms
- BLAST based identification of virulence genes
Download VFDB core dataset (includes genes associated with experimentally verified VFs only) and convert into BLAST database.
makeblastdb -in VFDB_db -dbtype 'nucl' -out <db_name>
Run BLASTn for clustering results against VFDB database.
blastn -db <db_name> -query <cluster> -out <result> -max_hsps 1 -max_target_seqs 1 -outfmt "6 qseqid length qstart qend sstart send evalue bitscore stitle" -perc_identity 100 -num_threads 5
CARD
- Comprehensive Antibiotic Resistance Database
- Provides data, models, and algorithms relating to the molecular basis of antimicrobial resistance
- Can be used for the analysis of genome sequences using the Resistance Gene Identifier
Install the RGI software using conda & download CARD database:
wget https://card.mcmaster.ca/latest/data tar -xvf data ./card.json
Build a local database:
rgi load --card_json <path to card.json> --local
Run RGI main with protein sequences:
rgi main -i <path to cluster.faa> -o <output_file_name> -t protein --local
Create a tab-delimited file from rgi results:
rgi tab -i <path to output_file_name.json>
MicrobesOnline
Predicts whether pairs of adjacent genes that are on the same strand are in the same operon, based on the intergenic distance between them, whether orthologs of the genes are near each other in other genomes, and their predicted functions.
- Download all the operon tables for Campylobacter jejuni.
- Use GID to retrieve the protein reference sequences.
- Create a database using BLAST:
makeblastdb -in <fasta file > -dbtype prot -out <database>
- Query the clustered sequences with the reference protein database:
blastp -query cdhit/faa_rep_seq.faa -db tmp/db_operon -evalue 0.01 -max_target_seqs 1 -max_hsps 1 -outfmt 6 -out tmp/hits_0.01.txt -num_threads 5
Ab Initio Methods
Categories
Transmembrane Proteins (Cell Membrane and Outer Membrane):
- Bacteria have the ability to export effector proteins in membranes of eukaryotic host
- Integral membrane protein that function as gates or docking sites that allow or prevent the entry or exit of materials across the cell membrane
Signal Peptides:
- Guide secretory proteins to find their correct locations outside the cell membrane for signal transduction
CRISPR:
- Provides immunity to the bacteria against Bacteriophages
- Contributes to the Virulence and Pathogenicity of the bacteria
TMHMM2
- Uses Hidden Markov Model approach
- Outputs the number of transmembrane helices predicted, the position of each residue with respect to the cell membrane (outside/inside the cell, transmembrane segment), and optional graphical output visualizing the predicted helix
Input a multifasta file into TMHMM.
tmhmm <input multifasta file> > <output file>
PilerCR
- Rapid identification and classification of CRISPR repeats
- High sensitivity and high specificity
Input a multifasta file into PileCR.
pilercr –in <input multifasta file> -out <output file> -noinfo –quiet
SignalP
- Uses a deep neural network algorithm
- Predicts presence of signal peptides and location of cleavage sites
- Does not determine lipoproteins
Input a fasta file into SignalP.
signalp –fasta <input_sequence_file> -org gram- -format short –gff3
Initial Pipeline
Results
Clustering
Ab Initio
Transmembrane Proteins
- Minimum = 810
- Maximum = 1,049
- Average = 928.8
Signal Peptides
- Minimum = 71
- Maximum = 99
- Average = 94.3
Lipoproteins
- Minimum = 43
- Maximum = 67
- Average = 52.8
CRISPR
- Minimum = 0
- Maximum = 2
- Average = 0.8
Homology
Antibiotic Resistance
- Minimum = 1
- Maximum = 10
- Average = 3.8
VFDB Ouput
Virulence Factors
- Total = 132
- Minimum = 1
- Maximum = 13
- Average = 3.1
Operons
- Minimum = 505
- Maximum = 856
- Average = 678.1
eggNog-mapper Ouput
EggNog-mapper provides useful functional annotations such as seed eggNOG ortholog, seed ortholog evalue/score, predicted taxonomic group, predicted protein name, GO terms, KEGG ko, KEGG Pathways, eggnog OG, COG Functional Category, free text descriptions, and many more. The descriptions of the COG Functional Categories are shown below.
eggNog-mapper Results
- Total = 44,098
- Minimum = 669
- Maximum = 1087
- Average = 881.9
Final Pipeline
References
Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1899501/
Genomic Characterization of Campylobacter jejuni Strain M1 . https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2928727/
A Genome-Wide Association Study to Identify Diagnostic Markers for Human Pathogenic Campylobacter jejuni Strains https://www.frontiersin.org/articles/10.3389/fmicb.2017.01224/full
Campylobacter jejuni transcriptional and genetic adaptation during human infection https://centerforimmunizationresearch.org/wp-content/uploads/2018/08/Campylobacter-jejuni-transcriptional-and-genetic-adaptation-during-human-infection.pdf
A proteome-wide protein interaction map for Campylobacter jejuni https://link.springer.com/article/10.1186/gb-2007-8-7-r130
Li, Weizhong & Fu, Limin & Wu, Sitao & Wooley, John. (2012). Ultrafast clustering algorithms for metagenomic sequence analysis. Briefings in bioinformatics. 13. 10.1093/bib/bbs035.
Xavier, Basil Britto et al. “Consolidating and Exploring Antibiotic Resistance Gene Data Resources.” Journal of clinical microbiology vol. 54,4 (2016): 851-9. doi:10.1128/JCM.02717-15
Huerta-Cepas, Jaime & Forslund, Sofia & Coelho, Luis Pedro & Szklarczyk, Damian & Jensen, Lars & von Mering, Christian & Bork, Peer. (2017). Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Molecular Biology and Evolution. 34. 10.1093/molbev/msx148.
Alcock, Brian P et al. "CARD 2020: Antibiotic Resistome Surveillance With The Comprehensive Antibiotic Resistance Database". Nucleic Acids Research, 2019. Oxford University Press (OUP), doi:10.1093/nar/gkz935
M. N. Price, E. J. Alm, and A. P. Arkin (2005). Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication.. Nucleic Acids Research 33:3224-3234.
Marasini, Daya et al. “Phylogenetic Relatedness Among Plasmids Harbored by Campylobacter jejuni and Campylobacter coli Isolated From Retail Meats.” Frontiers in microbiology vol. 9 2167. 12 Sep. 2018, doi:10.3389/fmicb.2018.02167
Carattoli, Alessandra et al. “In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing.” Antimicrobial agents and chemotherapy vol. 58,7 (2014): 3895-903. doi:10.1128/AAC.02412-14
Marasini, Daya et al. “Phylogenetic Relatedness Among Plasmids Harbored by Campylobacter jejuni and Campylobacter coli Isolated From Retail Meats.” Frontiers in microbiology vol. 9 2167. 12 Sep. 2018, doi:10.3389/fmicb.2018.02167
Carattoli, Alessandra et al. “In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing.” Antimicrobial agents and chemotherapy vol. 58,7 (2014): 3895-903. doi:10.1128/AAC.02412-14
Roosaare M, Puustusmaa M, Möls M, Vaher M, Remm M. 2018. PlasmidSeeker: identification of known plasmids from bacterial whole genome sequencing reads. PeerJ6:e4588 https://doi.org/10.7717/peerj.4588
Weizhong Li, Lukasz Jaroszewski, Adam Godzik, Clustering of highly homologous sequences to reduce the size of large protein databases , Bioinformatics, Volume 17, Issue 3, March 2001, Pages 282–283, https://doi.org/10.1093/bioinformatics/17.3.282
Steinegger, M., Söding, J. Clustering huge protein sequence sets in linear time. Nat Commun 9, 2542 (2018). https://doi.org/10.1038/s41467-018-04964-5
Benjamin T James, Brian B Luczak, Hani Z Girgis, MeShClust: an intelligent tool for clustering DNA sequences, Nucleic Acids Research, Volume 46, Issue 14, 21 August 2018, Page e83, https://doi.org/10.1093/nar/gky315
Robert C. Edgar, Search and clustering orders of magnitude faster than BLAST, Bioinformatics, Volume 26, Issue 19, 1 October 2010, Pages 2460–2461, https://doi.org/10.1093/bioinformatics/btq461
Soto SM. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence. 2013;4(3):223–229. doi:10.4161/viru.23724